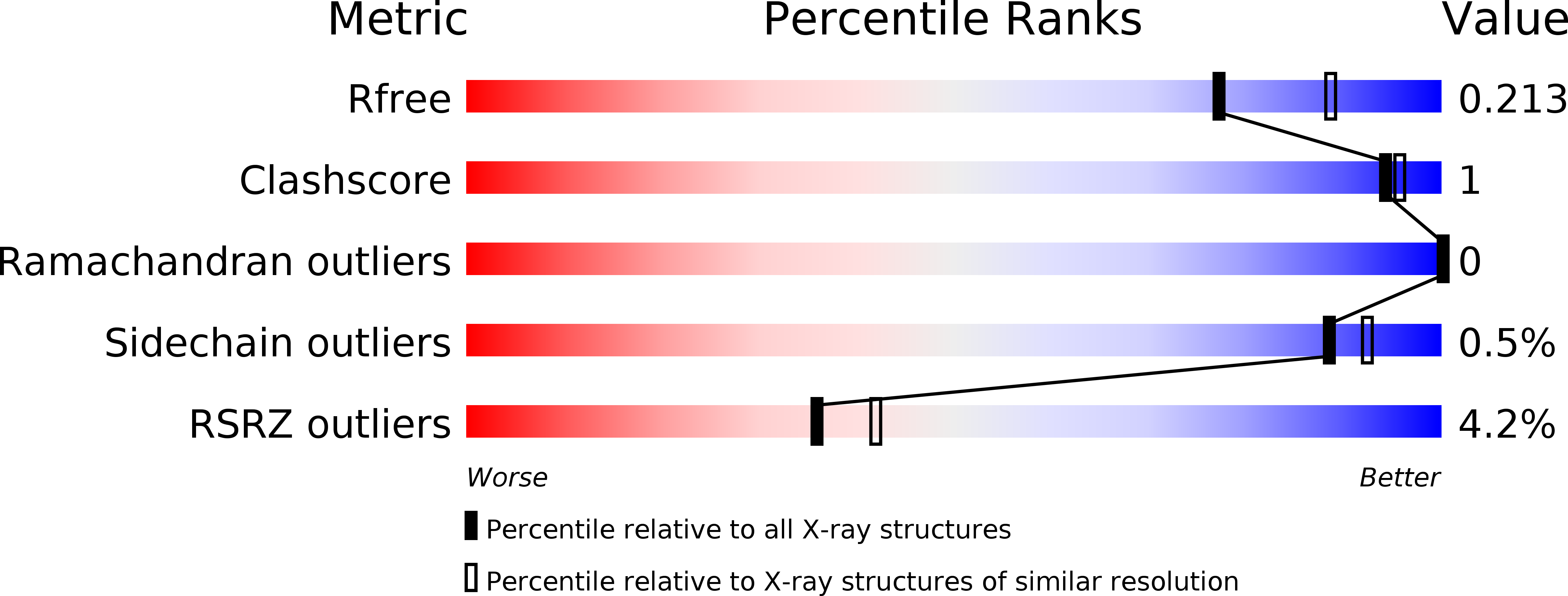
Non-Heme Monooxygenase ThoJ Catalyzes Thioholgamide beta-Hydroxylation.
Sikandar, A., Lopatniuk, M., Luzhetskyy, A., Koehnke, J.(2020) ACS Chem Biol 15: 2815-2819
- PubMed: 32965102
- DOI: https://doi.org/10.1021/acschembio.0c00637
- Primary Citation of Related Structures:
6ZYK, 6ZYL - PubMed Abstract:
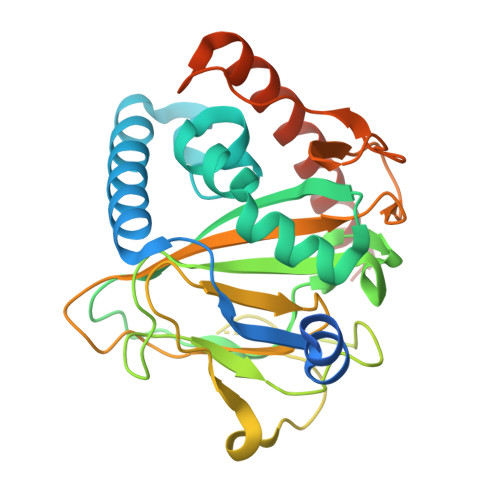
Thioviridamide-like compounds, including thioholgamides, are ribosomally synthesized and post-translationally modified peptide natural products with potent anticancer cell activity and an unprecedented structure. Very little is known about their biosynthesis, and we were intrigued by the β-hydroxy-N1, N3-dimethylhistidinium moiety found in these compounds. Here we report the construction of a heterologous host capable of producing thioholgamide with a 15-fold increased yield compared to the wild-type strain. A knockout of thoJ , encoding a predicted nonheme monooxygenase, shows that ThoJ is essential for thioholgamide β-hydroxylation. The crystal structure of ThoJ exhibits a typical mono/dioxygenase fold with conserved key active-site residues. Yet, ThoJ possesses a very large substrate binding pocket that appears suitable to receive a cyclic thioholgamide intermediate for hydroxylation. The improved production of the heterologous host will enable the dissection of the individual biosynthetic steps involved in biosynthesis of this exciting RiPP family.
Organizational Affiliation:
Workgroup Structural Biology of Biosynthetic Enzymes, Helmholtz Institute for Pharmaceutical Research Saarland (HIPS), Helmholtz Centre for Infection Research (HZI), Saarland University, Campus Geb. E8.1, 66123 Saarbrücken, Germany.