Discovery of ANT3310 , a Novel Broad-Spectrum Serine beta-Lactamase Inhibitor of the Diazabicyclooctane Class, Which Strongly Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacterales and Acinetobacter baumannii.
Davies, D.T., Leiris, S., Zalacain, M., Sprynski, N., Castandet, J., Bousquet, J., Lozano, C., Llanos, A., Alibaud, L., Vasa, S., Pattipati, R., Valige, R., Kummari, B., Pothukanuri, S., De Piano, C., Morrissey, I., Holden, K., Warn, P., Marcoccia, F., Benvenuti, M., Pozzi, C., Tassone, G., Mangani, S., Docquier, J.D., Pallin, D., Elliot, R., Lemonnier, M., Everett, M.(2020) J Med Chem 63: 15802-15820
- PubMed: 33306385
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01535
- Primary Citation of Related Structures:
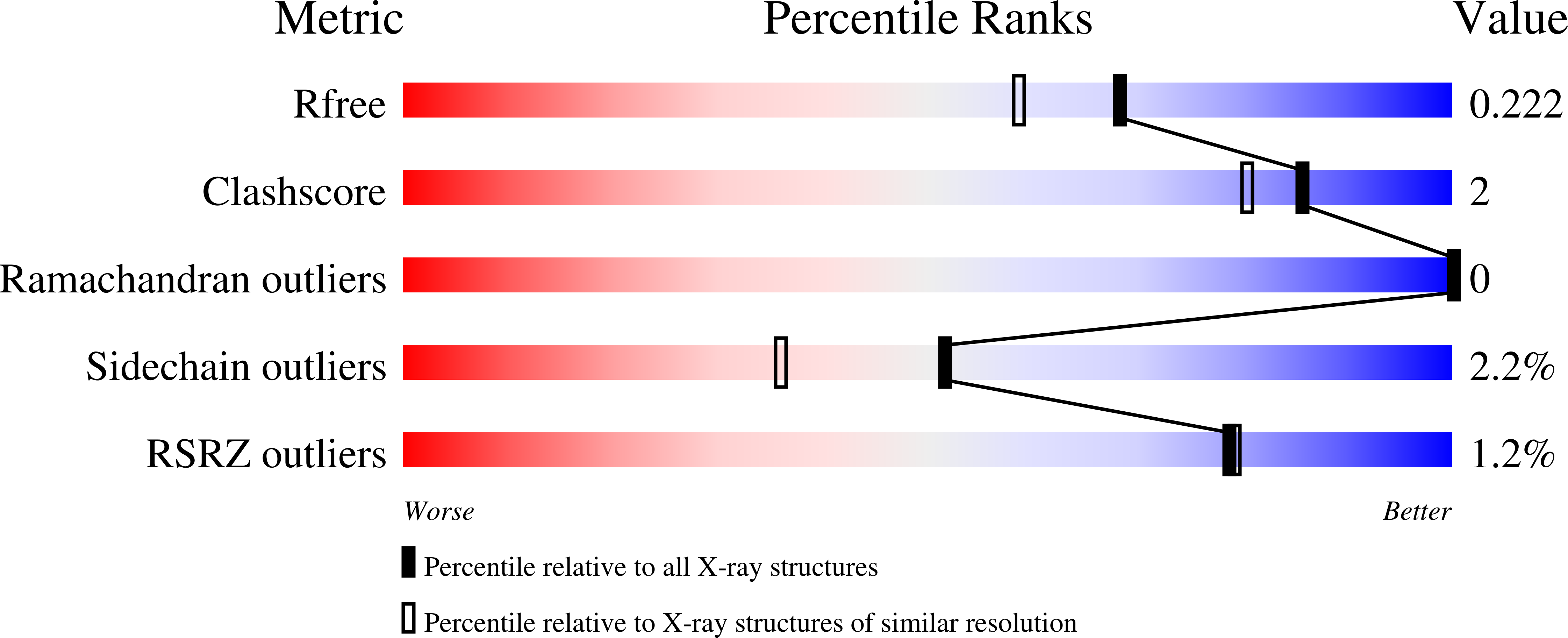
6ZXI - PubMed Abstract:
The diazabicyclooctanes (DBOs) are a class of serine β-lactamase (SBL) inhibitors that use a strained urea moiety as the warhead to react with the active serine residue in the active site of SBLs. The first in-class drug, avibactam, as well as several other recently approved DBOs (e.g., relebactam) or those in clinical development (e.g., nacubactam and zidebactam) potentiate activity of β-lactam antibiotics, to various extents, against carbapenem-resistant Enterobacterales (CRE) carrying class A, C, and D SBLs; however, none of these are able to rescue the activity of β-lactam antibiotics against carbapenem-resistant Acinetobacter baumannii (CRAB), a WHO "critical priority pathogen" producing class D OXA-type SBLs. Herein, we describe the chemical optimization and resulting structure-activity relationship, leading to the discovery of a novel DBO, ANT3310 , which uniquely has a fluorine atom replacing the carboxamide and stands apart from the current DBOs in restoring carbapenem activity against OXA-CRAB as well as SBL-carrying CRE pathogens.
Organizational Affiliation:
Antabio SAS, 436 rue Pierre et Marie Curie, 31670 Labège, France.