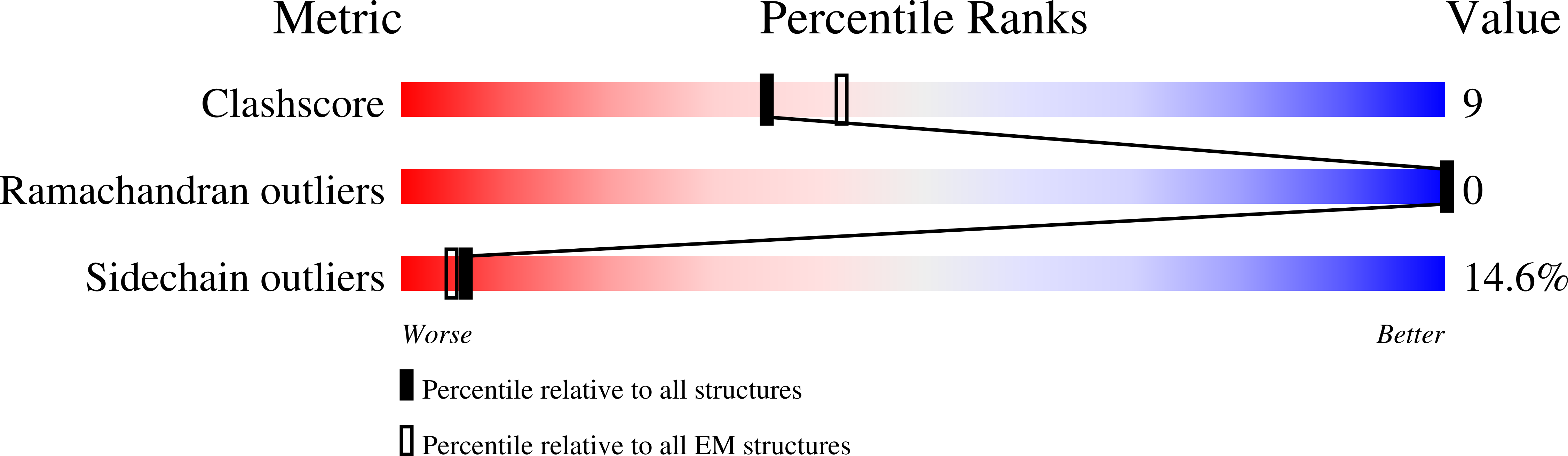Folding of an Intrinsically Disordered Iron-Binding Peptide in Response to Sedimentation Revealed by Cryo-EM.
Davidov, G., Abelya, G., Zalk, R., Izbicki, B., Shaibi, S., Spektor, L., Shagidov, D., Meyron-Holtz, E.G., Zarivach, R., Frank, G.A.(2020) J Am Chem Soc 142: 19551-19557
- PubMed: 33166133
- DOI: https://doi.org/10.1021/jacs.0c07565
- Primary Citation of Related Structures:
6Z3D, 6ZH5, 6ZLG, 6ZLQ - PubMed Abstract:
Biomineralization is mediated by specialized proteins that guide and control mineral sedimentation. In many cases, the active regions of these biomineralization proteins are intrinsically disordered. High-resolution structures of these proteins while they interact with minerals are essential for understanding biomineralization processes and the function of intrinsically disordered proteins (IDPs). Here we used the cavity of ferritin as a nanoreactor where the interaction between M6A, an intrinsically disordered iron-binding domain, and an iron oxide particle was visualized at high resolution by cryo-EM. Taking advantage of the differences in the electron-dose sensitivity of the protein and the iron oxide particles, we developed a method to determine the irregular shape of the particles found in our density maps. We found that the folding of M6A correlates with the detection of mineral particles in its vicinity. M6A interacts with the iron oxide particles through its C-terminal side, resulting in the stabilization of a helix at its N-terminal side. The stabilization of the helix at a region that is not in direct contact with the iron oxide particle demonstrates the ability of IDPs to respond to signals from their surroundings by conformational changes. These findings provide the first glimpse toward the long-suspected mechanism for biomineralization protein control over mineral microstructure, where unstructured regions of these proteins become more ordered in response to their interaction with the nascent mineral particles.
Organizational Affiliation:
Department of Life Sciences, Ben-Gurion University of the Negev, Beer Sheva 8410501, Israel.