[ b ]-Annulated Halogen-Substituted Indoles as Potential DYRK1A Inhibitors.
Lechner, C., Flasshoff, M., Falke, H., Preu, L., Loaec, N., Meijer, L., Knapp, S., Chaikuad, A., Kunick, C.(2019) Molecules 24
- PubMed: 31766108
- DOI: https://doi.org/10.3390/molecules24224090
- Primary Citation of Related Structures:
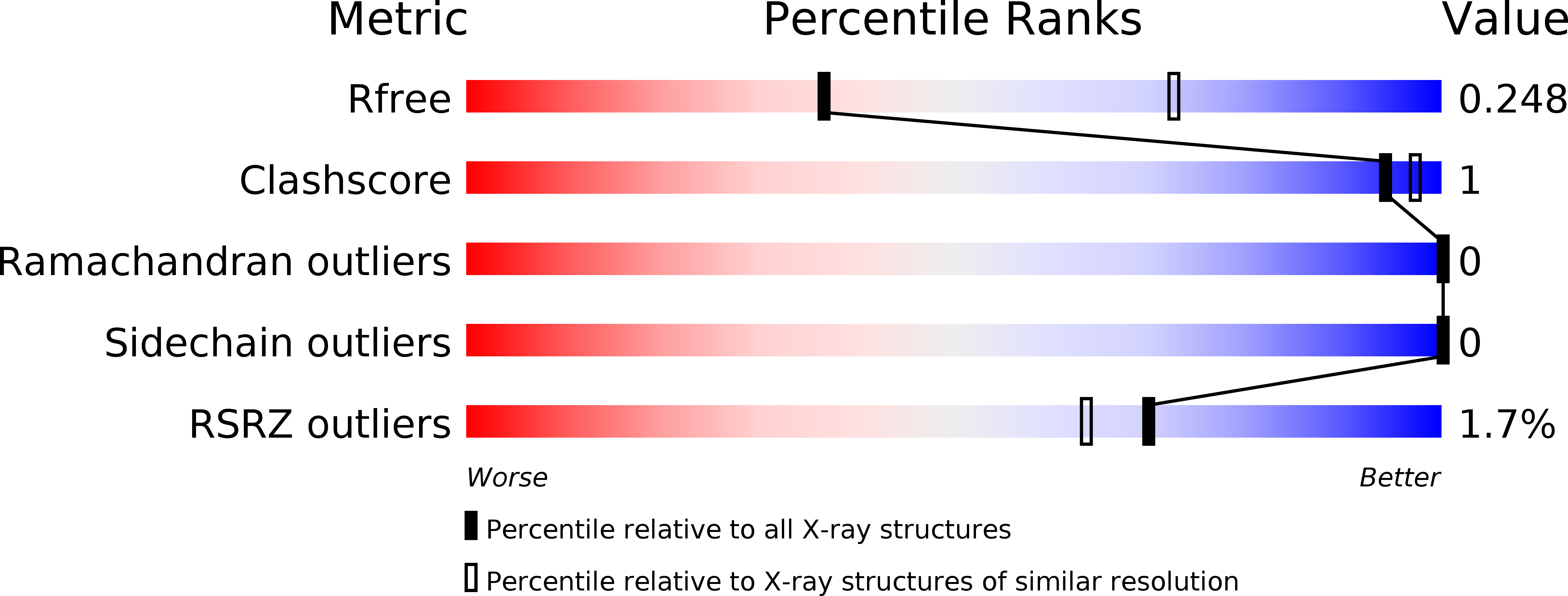
6T6A - PubMed Abstract:
Since hyperactivity of the protein kinase DYRK1A is linked to several neurodegenerative disorders, DYRK1A inhibitors have been suggested as potential therapeutics for Down syndrome and Alzheimer's disease. Most published inhibitors to date suffer from low selectivity against related kinases or from unfavorable physicochemical properties. In order to identify DYRK1A inhibitors with improved properties, a series of new chemicals based on [ b ]-annulated halogenated indoles were designed, synthesized, and evaluated for biological activity. Analysis of crystal structures revealed a typical type-I binding mode of the new inhibitor 4-chlorocyclohepta[ b ]indol-10(5 H )-one in DYRK1A, exploiting mainly shape complementarity for tight binding. Conversion of the DYRK1A inhibitor 8-chloro-1,2,3,9-tetrahydro-4 H -carbazol-4-one into a corresponding Mannich base hydrochloride improved the aqueous solubility but abrogated kinase inhibitory activity.
Organizational Affiliation:
Institut für Medizinische und Pharmazeutische Chemie, Technische Universität Braunschweig, Beethovenstraße 55, 38106 Braunschweig, Germany.