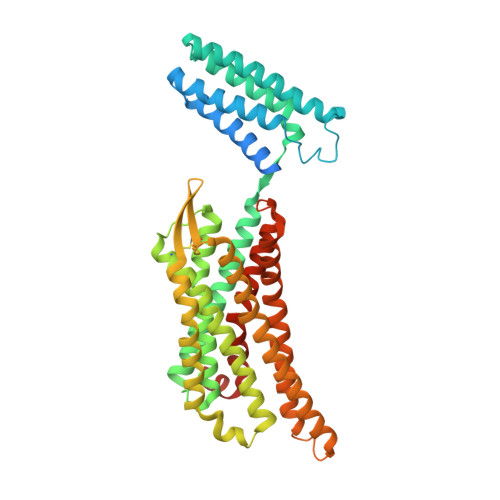
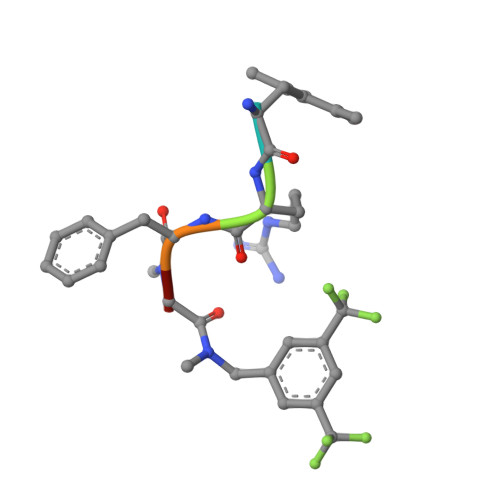
Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Claff, T., Yu, J., Blais, V., Patel, N., Martin, C., Wu, L., Han, G.W., Holleran, B.J., Van der Poorten, O., White, K.L., Hanson, M.A., Sarret, P., Gendron, L., Cherezov, V., Katritch, V., Ballet, S., Liu, Z.J., Muller, C.E., Stevens, R.C.(2019) Sci Adv 5: eaax9115-eaax9115
- PubMed: 31807708
- DOI: https://doi.org/10.1126/sciadv.aax9115
- Primary Citation of Related Structures:
6PT2, 6PT3 - PubMed Abstract:
Selective activation of the δ-opioid receptor (DOP) has great potential for the treatment of chronic pain, benefitting from ancillary anxiolytic and antidepressant-like effects. Moreover, DOP agonists show reduced adverse effects as compared to μ-opioid receptor (MOP) agonists that are in the spotlight of the current "opioid crisis." Here, we report the first crystal structures of the DOP in an activated state, in complex with two relevant and structurally diverse agonists: the potent opioid agonist peptide KGCHM07 and the small-molecule agonist DPI-287 at 2.8 and 3.3 Å resolution, respectively. Our study identifies key determinants for agonist recognition, receptor activation, and DOP selectivity, revealing crucial differences between both agonist scaffolds. Our findings provide the first investigation into atomic-scale agonist binding at the DOP, supported by site-directed mutagenesis and pharmacological characterization. These structures will underpin the future structure-based development of DOP agonists for an improved pain treatment with fewer adverse effects.
Organizational Affiliation:
iHuman Institute, ShanghaiTech University, Ren Building, 393 Middle Huaxia Rd, Pudong, Shanghai 201210, China.