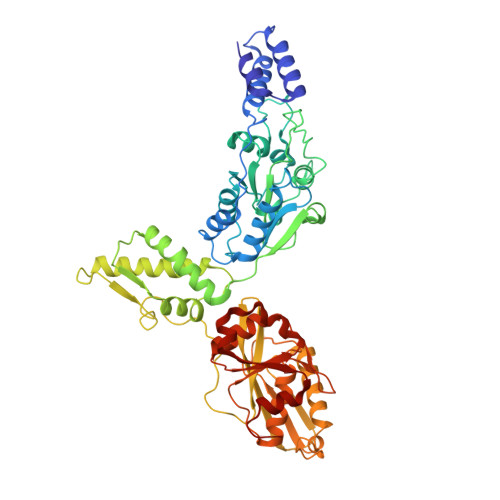
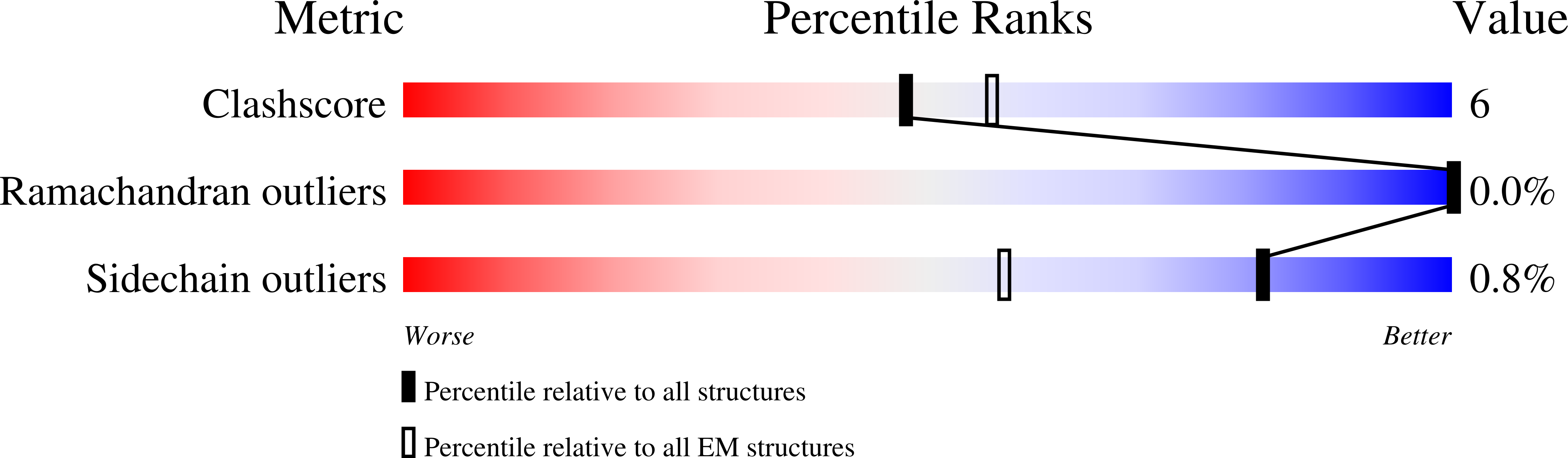Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Shin, M., Puchades, C., Asmita, A., Puri, N., Adjei, E., Wiseman, R.L., Karzai, A.W., Lander, G.C.(2020) Sci Adv 6: eaba8404-eaba8404
- PubMed: 32490208
- DOI: https://doi.org/10.1126/sciadv.aba8404
- Primary Citation of Related Structures:
6ON2, 6V11 - PubMed Abstract:
Substrate-bound structures of AAA+ protein translocases reveal a conserved asymmetric spiral staircase architecture wherein a sequential ATP hydrolysis cycle drives hand-over-hand substrate translocation. However, this configuration is unlikely to represent the full conformational landscape of these enzymes, as biochemical studies suggest distinct conformational states depending on the presence or absence of substrate. Here, we used cryo-electron microscopy to determine structures of the Yersinia pestis Lon AAA+ protease in the absence and presence of substrate, uncovering the mechanistic basis for two distinct operational modes. In the absence of substrate, Lon adopts a left-handed, "open" spiral organization with autoinhibited proteolytic active sites. Upon the addition of substrate, Lon undergoes a reorganization to assemble an enzymatically active, right-handed "closed" conformer with active protease sites. These findings define the mechanistic principles underlying the operational plasticity required for processing diverse protein substrates.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.