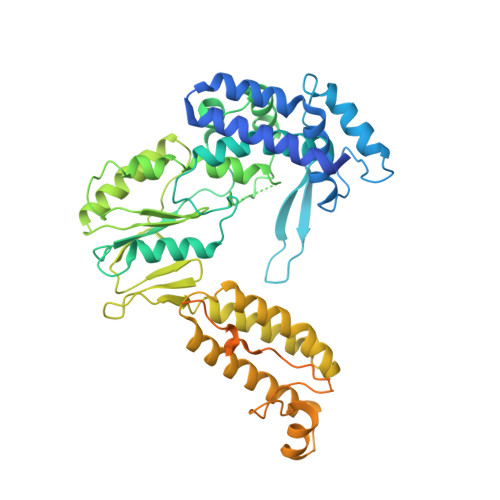
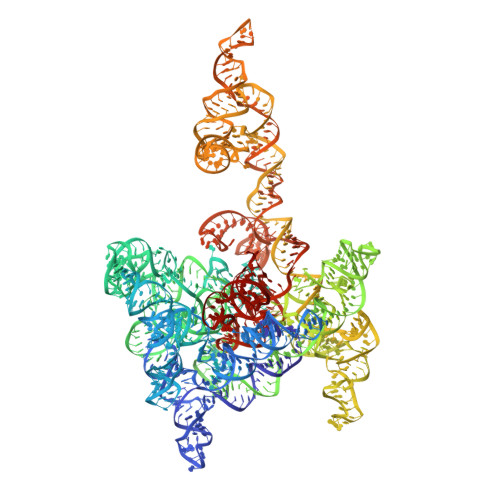
Cryo-EM Structures of a Group II Intron Reverse Splicing into DNA.
Haack, D.B., Yan, X., Zhang, C., Hingey, J., Lyumkis, D., Baker, T.S., Toor, N.(2019) Cell 178: 612-623.e12
- PubMed: 31348888
- DOI: https://doi.org/10.1016/j.cell.2019.06.035
- Primary Citation of Related Structures:
6ME0, 6MEC - PubMed Abstract:
Group II introns are a class of retroelements that invade DNA through a copy-and-paste mechanism known as retrotransposition. Their coordinated activities occur within a complex that includes a maturase protein, which promotes splicing through an unknown mechanism. The mechanism of splice site exchange within the RNA active site during catalysis also remains unclear. We determined two cryo-EM structures at 3.6-Å resolution of a group II intron reverse splicing into DNA. These structures reveal that the branch-site domain VI helix swings 90°, enabling substrate exchange during DNA integration. The maturase assists catalysis through a transient RNA-protein contact with domain VI that positions the branch-site adenosine for lariat formation during forward splicing. These findings provide the first direct evidence of the role the maturase plays during group II intron catalysis. The domain VI dynamics closely parallel spliceosomal branch-site helix movement and provide strong evidence for a retroelement origin of the spliceosome.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093, USA.