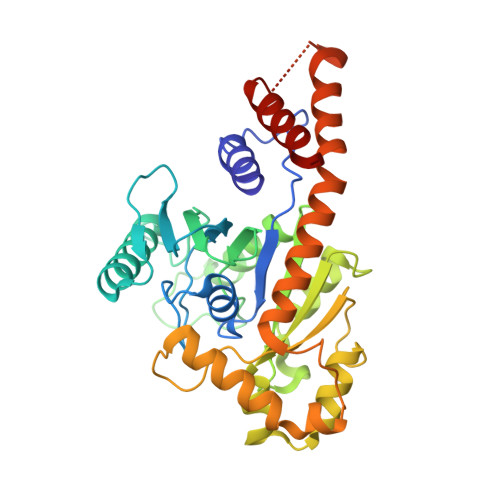
Structural insights into G domain dimerization and pathogenic mutation of OPA1.
Yu, C., Zhao, J., Yan, L., Qi, Y., Guo, X., Lou, Z., Hu, J., Rao, Z.(2020) J Cell Biol 219
- PubMed: 32379273
- DOI: https://doi.org/10.1083/jcb.201907098
- Primary Citation of Related Structures:
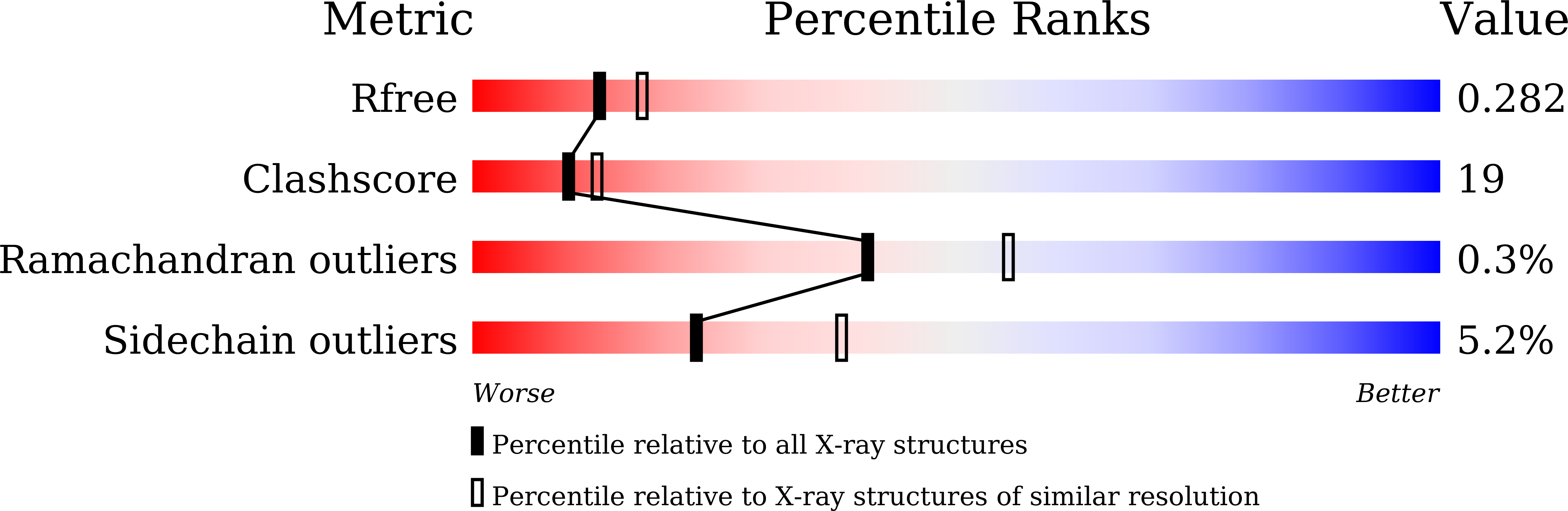
6JTG - PubMed Abstract:
The fusion of mammalian inner mitochondrial membranes (IMMs) is mediated by dynamin-like GTPase OPA1. Mutations in human OPA1 cause optic atrophy, but the molecular basis for membrane fusion and pathogenesis is not clear. Here, we determined the crystal structure of the minimal GTPase domain (MGD) of human OPA1. A three-helix bundle (HB) domain including two helices extending from the GTPase (G) domain and the last helix of OPA1 tightly associates with the G domain. In the presence of GDP and BeF3-, OPA1-MGD forms a dimer, the interface of which is critical for the maintenance of mitochondrial morphology. The catalytic core of OPA1 possesses unique features that are not present in other dynamin-like proteins. Biochemical experiments revealed that OPA1-MGD forms nucleotide-dependent dimers, which is important for membrane-stimulated GTP hydrolysis, and an N-terminal extension mediates nucleotide-independent dimerization that facilitates efficient membrane association. Our results suggest a multifaceted assembly of OPA1 and explain the effect of most OPA1 mutations on optic atrophy.
Organizational Affiliation:
College of Life Sciences and State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China.