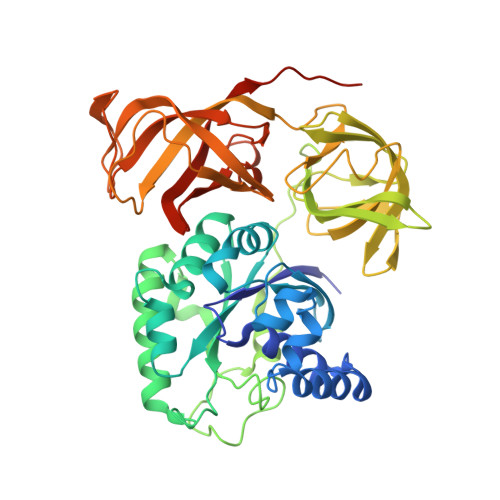
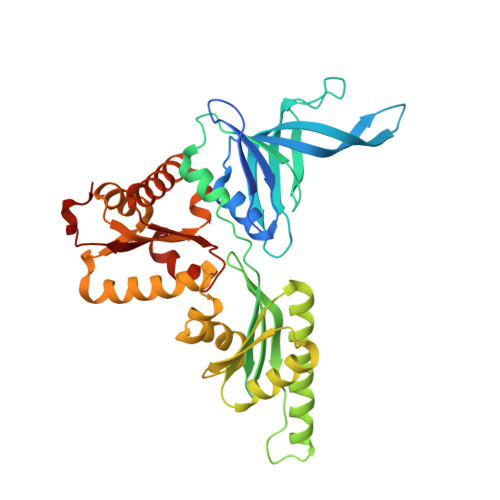
Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Maruyama, K., Imai, H., Kawamura, M., Ishino, S., Ishino, Y., Ito, K., Uchiumi, T.(2019) Sci Rep 9: 14761-14761
- PubMed: 31611569
- DOI: https://doi.org/10.1038/s41598-019-51266-x
- Primary Citation of Related Structures:
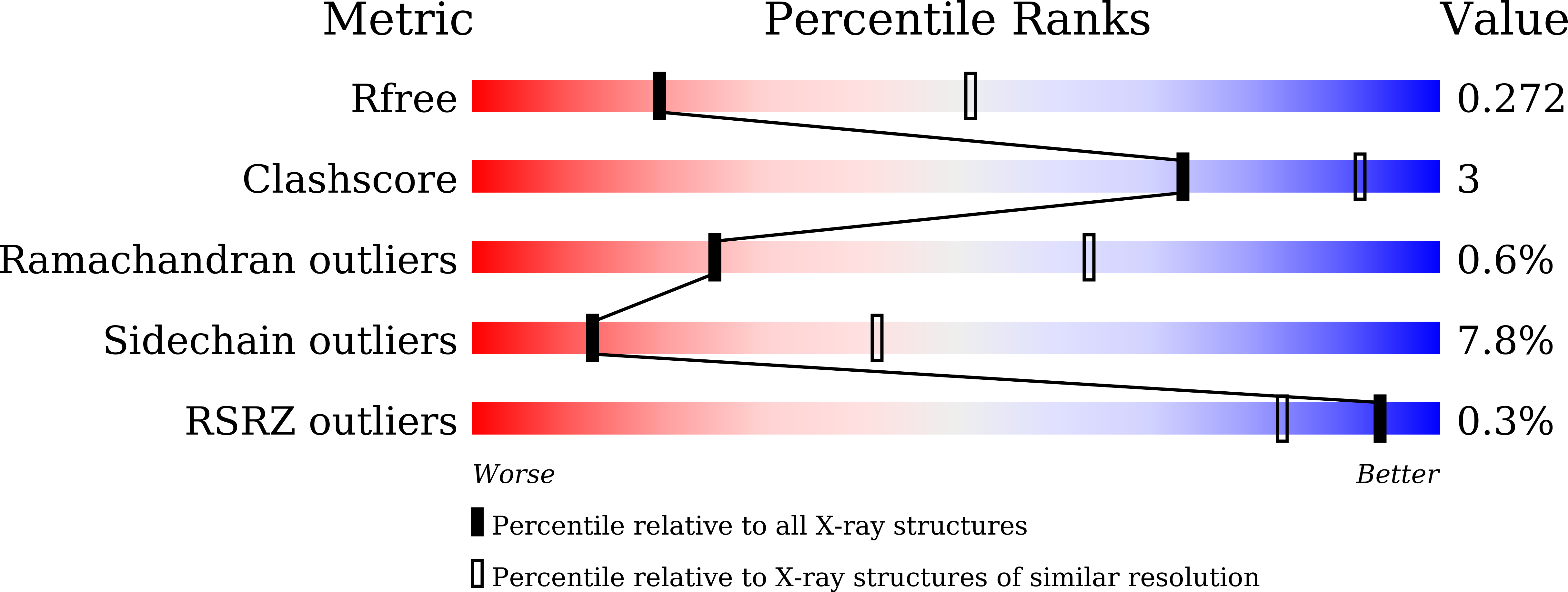
6JI2 - PubMed Abstract:
Translation elongation factor EF1A delivers aminoacyl-tRNA to the ribosome in a GTP-bound form, and is released from the ribosome in a GDP-bound form. This association/dissociation cycle proceeds efficiently via a marked conformational change in EF1A. EF1A function is dependent on the ribosomal "stalk" protein of the ribosomal large subunit, although the precise mechanism of action of the stalk on EF1A remains unclear. Here, we clarify the binding mode of archaeal stalk aP1 to GTP-bound aEF1A associated with aPelota. Intriguingly, the C-terminal domain (CTD) of aP1 binds to aEF1A•GTP with a similar affinity to aEF1A•GDP. We have also determined the crystal structure of the aP1-CTD•aEF1A•GTP•aPelota complex at 3.0 Å resolution. The structure shows that aP1-CTD binds to a space between domains 1 and 3 of aEF1A. Biochemical analyses show that this binding is crucial for protein synthesis. Comparison of the structures of aP1-CTD•aEF1A•GTP and aP1-CTD•aEF1A•GDP demonstrates that the binding mode of aP1 changes markedly upon a conformational switch between the GTP- and GDP-bound forms of aEF1A. Taking into account biochemical data, we infer that aP1 employs its structural flexibility to bind to aEF1A before and after GTP hydrolysis for efficient protein synthesis.
Organizational Affiliation:
Department of Biology, Faculty of Science, Niigata University, Ikarashi 2-8050, Niigata, 950-2181, Japan.