Structural basis for receptor recognition by Lujo virus.
Cohen-Dvashi, H., Kilimnik, I., Diskin, R.(2018) Nat Microbiol 3: 1153-1160
- PubMed: 30150732
- DOI: https://doi.org/10.1038/s41564-018-0224-5
- Primary Citation of Related Structures:
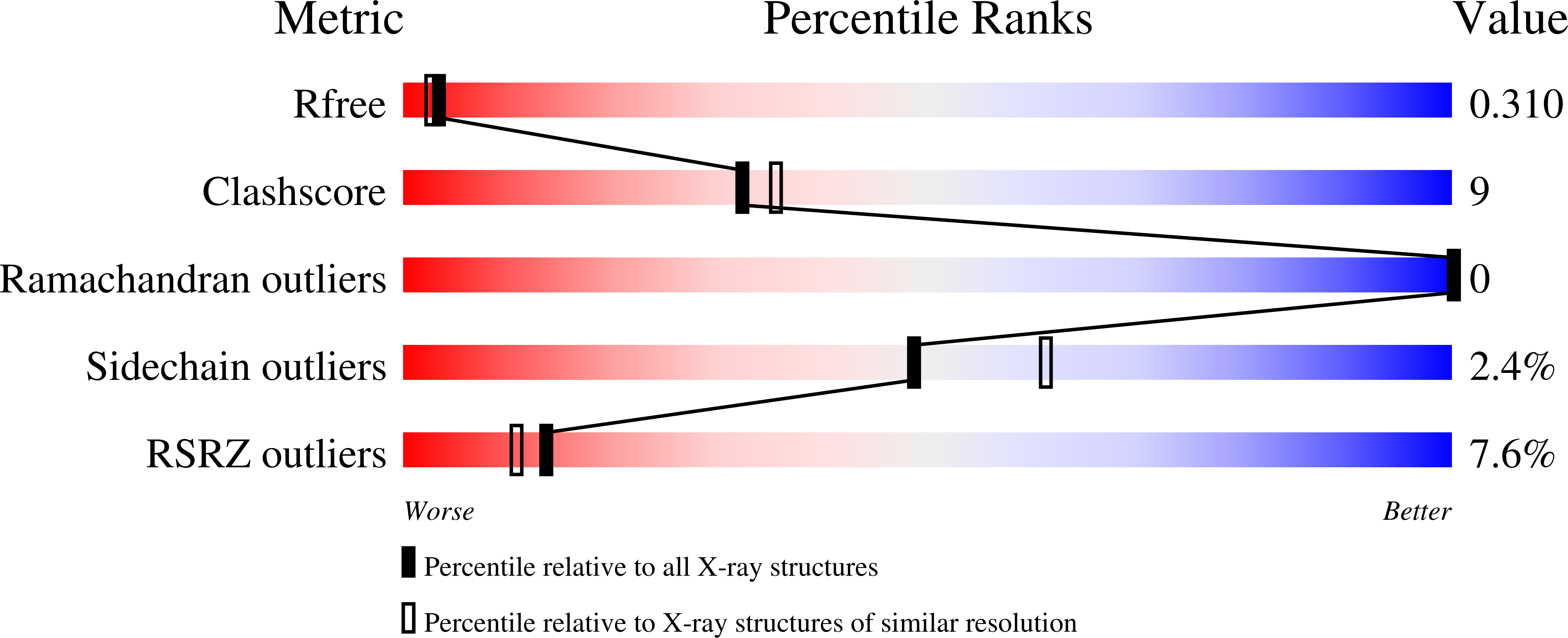
6GH8 - PubMed Abstract:
Lujo virus (LUJV) has emerged as a highly fatal human pathogen. Despite its membership among the Arenaviridae, LUJV does not classify with the known Old and New World groups of that viral family. Likewise, LUJV was recently found to use neuropilin-2 (NRP2) as a cellular receptor instead of the canonical receptors used by Old World and New World arenaviruses. The emergence of a deadly pathogen into human populations using an unprecedented entry route raises many questions regarding the mechanism of cell recognition. To provide the basis for combating LUJV in particular, and to increase our general understanding of the molecular changes that accompany an evolutionary switch to a new receptor for arenaviruses, we used X-ray crystallography to reveal how the GP1 receptor-binding domain of LUJV (LUJV GP1 ) recognizes NRP2. Structural data show that LUJV GP1 is more similar to Old World than to New World arenaviruses. Structural analysis supported by experimental validation further suggests that NRP2 recognition is metal-ion dependent and that the complete NRP2 binding site is formed in the context of the trimeric spike. Taken together, our data provide the mechanism for the cell attachment step of LUJV and present indispensable information for combating this phatogen.
Organizational Affiliation:
Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel.