Structural basis for anthrax toxin receptor 1 recognition by Seneca Valley Virus.
Jayawardena, N., Burga, L.N., Easingwood, R.A., Takizawa, Y., Wolf, M., Bostina, M.(2018) Proc Natl Acad Sci U S A 115: E10934-E10940
- PubMed: 30381454
- DOI: https://doi.org/10.1073/pnas.1810664115
- Primary Citation of Related Structures:
6CX1 - PubMed Abstract:
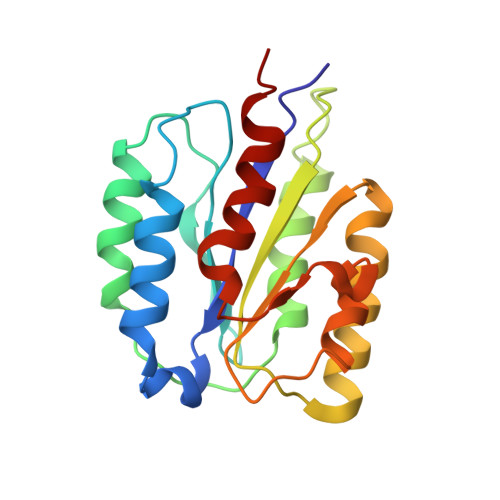
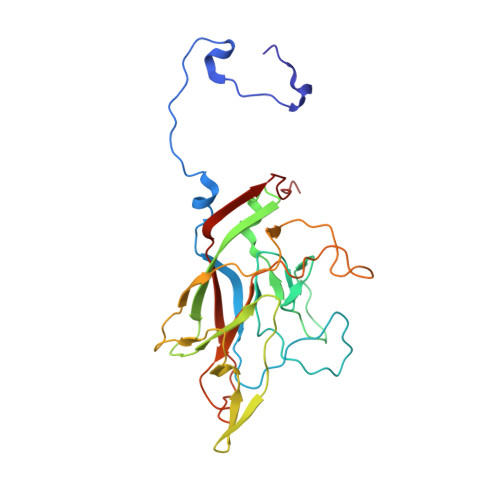
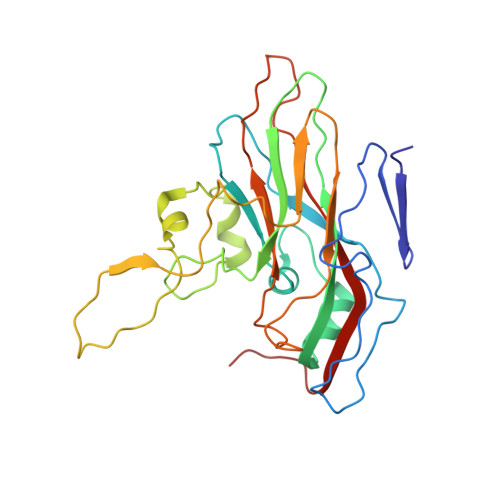
Recently, the use of oncolytic viruses in cancer therapy has become a realistic therapeutic option. Seneca Valley Virus (SVV) is a newly discovered picornavirus, which has earned a significant reputation as a potent oncolytic agent. Anthrax toxin receptor 1 (ANTXR1), one of the cellular receptors for the protective antigen secreted by Bacillus anthracis , has been identified as the high-affinity cellular receptor for SVV. Here, we report the structure of the SVV-ANTXR1 complex determined by single-particle cryo-electron microscopy analysis at near-atomic resolution. This is an example of a shared receptor structure between a mammalian virus and a bacterial toxin. Our structure shows that ANTXR1 decorates the outer surface of the SVV capsid and interacts with the surface-exposed BC loop and loop II of VP1, "the puff" of VP2 and "the knob" of VP3. Comparison of the receptor-bound capsid structure with the native capsid structure reveals that receptor binding induces minor conformational changes in SVV capsid structure, suggesting the role of ANTXR1 as an attachment receptor. Furthermore, our results demonstrate that the capsid footprint on the receptor is not conserved in anthrax toxin receptor 2 (ANTXR2), thereby providing a molecular mechanism for explaining the exquisite selectivity of SVV for ANTXR1.
Organizational Affiliation:
Department of Microbiology and Immunology, University of Otago, 9054 Dunedin, New Zealand.