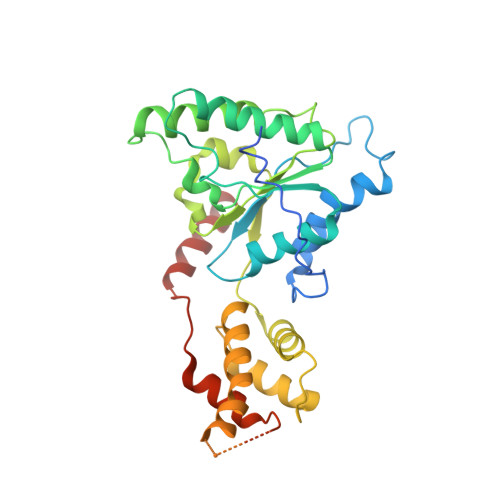
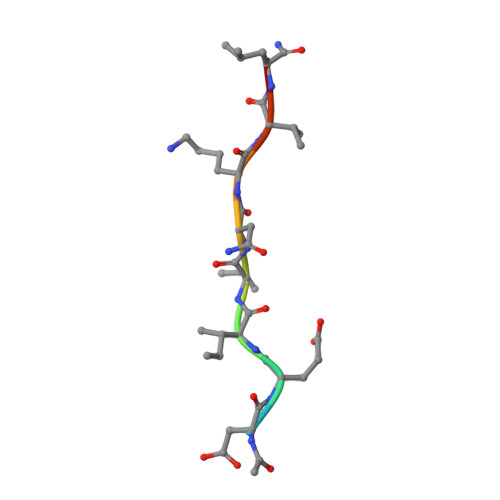
The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Han, H., Monroe, N., Sundquist, W.I., Shen, P.S., Hill, C.P.(2017) Elife 6
- PubMed: 29165244
- DOI: https://doi.org/10.7554/eLife.31324
- Primary Citation of Related Structures:
6AP1, 6BMF - PubMed Abstract:
The hexameric AAA ATPase Vps4 drives membrane fission by remodeling and disassembling ESCRT-III filaments. Building upon our earlier 4.3 Å resolution cryo-EM structure (Monroe et al., 2017), we now report a 3.2 Å structure of Vps4 bound to an ESCRT-III peptide substrate. The new structure reveals that the peptide approximates a β-strand conformation whose helical symmetry matches that of the five Vps4 subunits it contacts directly. Adjacent Vps4 subunits make equivalent interactions with successive substrate dipeptides through two distinct classes of side chain binding pockets formed primarily by Vps4 pore loop 1. These pockets accommodate a wide range of residues, while main chain hydrogen bonds may help dictate substrate-binding orientation. The structure supports a 'conveyor belt' model of translocation in which ATP binding allows a Vps4 subunit to join the growing end of the helix and engage the substrate, while hydrolysis and release promotes helix disassembly and substrate release at the lagging end.
Organizational Affiliation:
Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, United States.