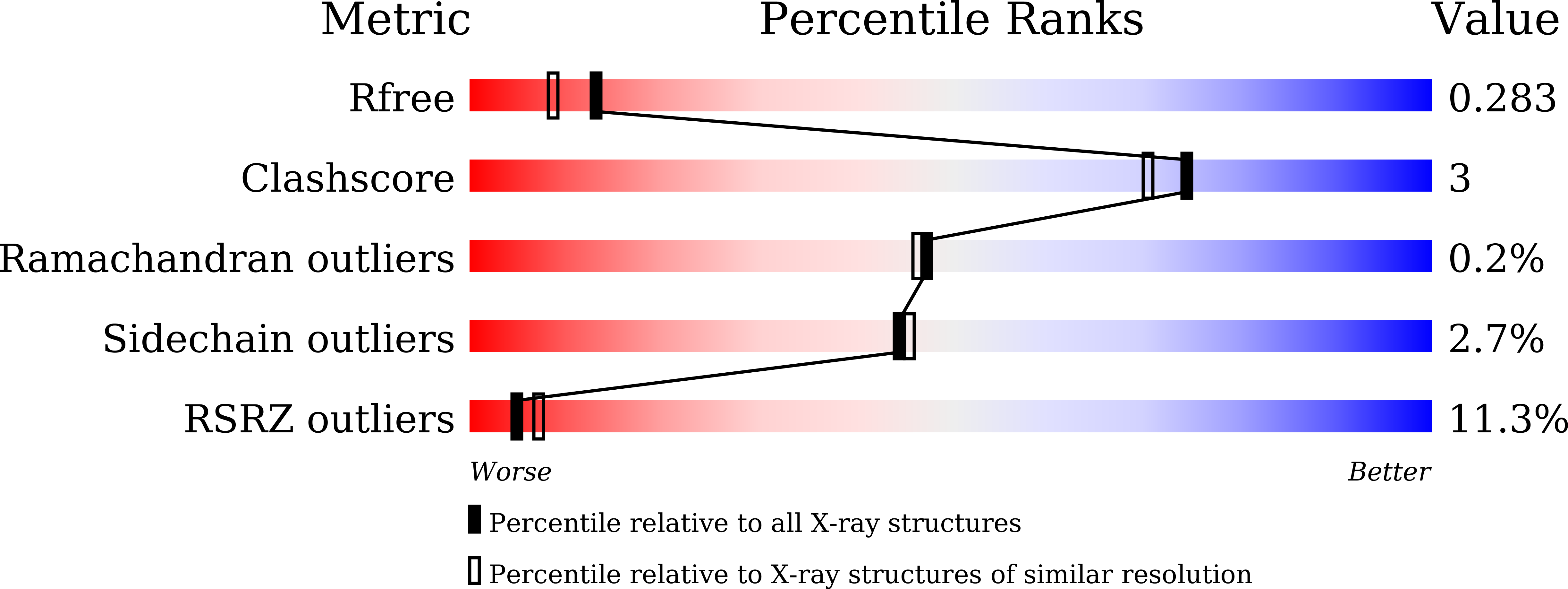
Discovery of DS42450411 as a potent orally active hepcidin production inhibitor: Design and optimization of novel 4-aminopyrimidine derivatives.
Fukuda, T., Ishiyama, T., Katagiri, T., Ueda, K., Muramatsu, S., Hashimoto, M., Aki, A., Baba, D., Watanabe, K., Tanaka, N.(2018) Bioorg Med Chem Lett 28: 3333-3337
- PubMed: 30217414
- DOI: https://doi.org/10.1016/j.bmcl.2018.09.010
- Primary Citation of Related Structures:
6A1F, 6A1G - PubMed Abstract:
Hepcidin has emerged as the central regulatory molecule in systemic iron homeostasis. The inhibition of hepcidin may be a favorable strategy for the treatment of anemia of chronic disease. Here, we have reported the design, synthesis, and structure-activity relationships (SAR) of a series of 4-aminopyrimidine compounds as inhibitors of hepcidin production. The optimization study of 1 led to the design of a potent and bioavailable inhibitor of hepcidin production, 34 (DS42450411), which showed serum hepcidin-lowering effects in a mouse model of interleukin-6-induced acute inflammation.
Organizational Affiliation:
Rare Disease Laboratories, Daiichi Sankyo Co., Ltd, 1-2-58 Hiromachi, Shinagawa-ku, Tokyo 140-8710, Japan. Electronic address: fukuda.takeshi.zv@daiichisankyo.co.jp.