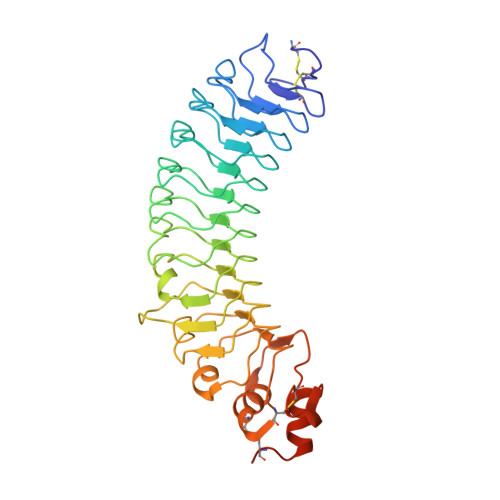
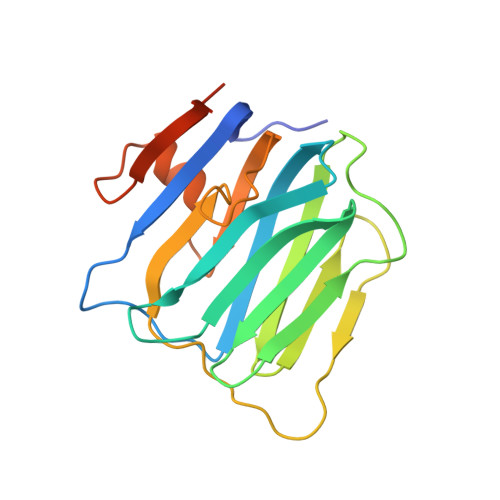
Structural insights into modulation and selectivity of transsynaptic neurexin-LRRTM interaction.
Yamagata, A., Goto-Ito, S., Sato, Y., Shiroshima, T., Maeda, A., Watanabe, M., Saitoh, T., Maenaka, K., Terada, T., Yoshida, T., Uemura, T., Fukai, S.(2018) Nat Commun 9: 3964-3964
- PubMed: 30262834
- DOI: https://doi.org/10.1038/s41467-018-06333-8
- Primary Citation of Related Structures:
5Z8X, 5Z8Y - PubMed Abstract:
Leucine-rich repeat transmembrane neuronal proteins (LRRTMs) function as postsynaptic organizers that induce excitatory synapses. Neurexins (Nrxns) and heparan sulfate proteoglycans have been identified as presynaptic ligands for LRRTMs. Specifically, LRRTM1 and LRRTM2 bind to the Nrxn splice variant lacking an insert at the splice site 4 (S4). Here, we report the crystal structure of the Nrxn1β-LRRTM2 complex at 3.4 Å resolution. The Nrxn1β-LRRTM2 interface involves Ca 2+ -mediated interactions and overlaps with the Nrxn-neuroligin interface. Together with structure-based mutational analyses at the molecular and cellular levels, the present structural analysis unveils the mechanism of selective binding between Nrxn and LRRTM1/2 and its modulation by the S4 insertion of Nrxn.
Organizational Affiliation:
Institute for Quantitative Biosciences, The University of Tokyo, Tokyo, 113-0032, Japan.