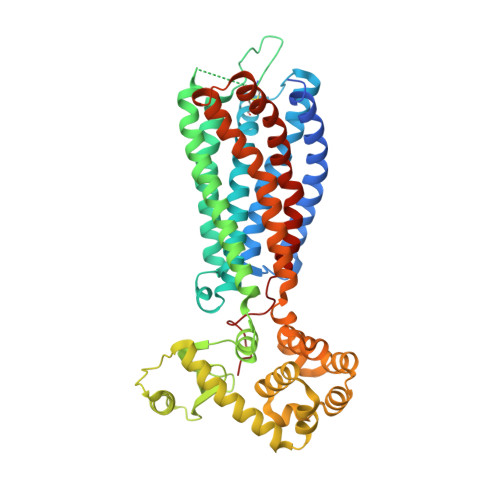
Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6
Taniguchi, R., Inoue, A., Sayama, M., Uwamizu, A., Yamashita, K., Hirata, K., Yoshida, M., Tanaka, Y., Kato, H.E., Nakada-Nakura, Y., Otani, Y., Nishizawa, T., Doi, T., Ohwada, T., Ishitani, R., Aoki, J., Nureki, O.(2017) Nature 548: 356-360
- PubMed: 28792932
- DOI: https://doi.org/10.1038/nature23448
- Primary Citation of Related Structures:
5XSZ - PubMed Abstract:
Lysophosphatidic acid (LPA) is a bioactive lipid composed of a phosphate group, a glycerol backbone, and a single acyl chain that varies in length and saturation. LPA activates six class A G-protein-coupled receptors to provoke various cellular reactions. Because LPA signalling has been implicated in cancer and fibrosis, the LPA receptors are regarded as promising drug targets. The six LPA receptors are subdivided into the endothelial differentiation gene (EDG) family (LPA 1 -LPA 3 ) and the phylogenetically distant non-EDG family (LPA 4 -LPA 6 ). The structure of LPA 1 has enhanced our understanding of the EDG family of LPA receptors. By contrast, the functional and pharmacological characteristics of the non-EDG family of LPA receptors have remained unknown, owing to the lack of structural information. Although the non-EDG LPA receptors share sequence similarity with the P2Y family of nucleotide receptors, the LPA recognition mechanism cannot be deduced from the P2Y 1 and P2Y 12 structures because of the large differences in the chemical structures of their ligands. Here we determine the 3.2 Å crystal structure of LPA 6 , the gene deletion of which is responsible for congenital hair loss, to clarify the ligand recognition mechanism of the non-EDG family of LPA receptors. Notably, the ligand-binding pocket of LPA 6 is laterally open towards the membrane, and the acyl chain of the lipid used for the crystallization is bound within this pocket, indicating the binding mode of the LPA acyl chain. Docking and mutagenesis analyses also indicated that the conserved positively charged residues within the central cavity recognize the phosphate head group of LPA by inducing an inward shift of transmembrane helices 6 and 7, suggesting that the receptor activation is triggered by this conformational rearrangement.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku, Tokyo 113-0032, Japan.