A Structural Rationale for N-Methylbicuculline Acting as a Promiscuous Competitive Antagonist of Inhibitory Pentameric Ligand-Gated Ion Channels.
Jones, M.J., Dawson, A., Hales, T.G., Hunter, W.N.(2020) Chembiochem 21: 1526-1533
- PubMed: 31859406
- DOI: https://doi.org/10.1002/cbic.201900680
- Primary Citation of Related Structures:
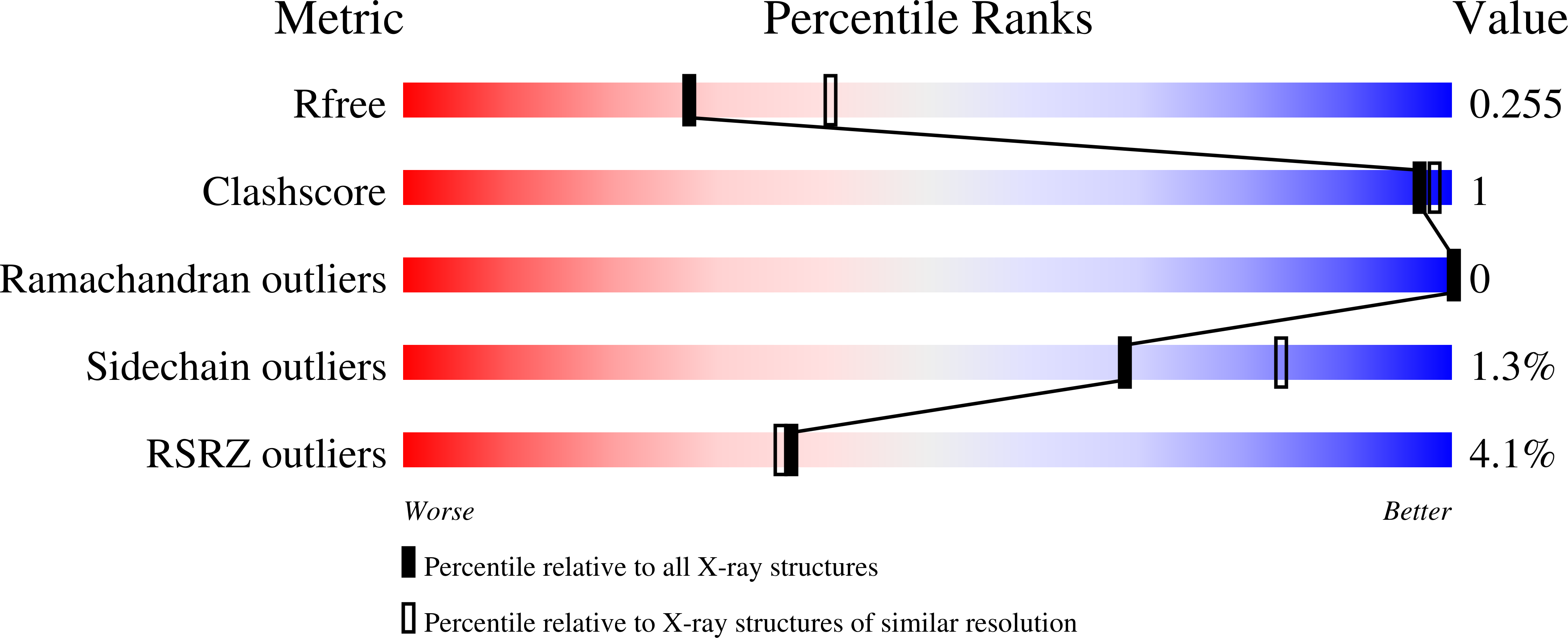
5OBH - PubMed Abstract:
Bicuculline, a valued chemical tool in neurosciences research, is a competitive antagonist of specific GABA A receptors and affects other pentameric ligand-gated ion channels including the glycine, nicotinic acetylcholine and 5-hydroxytryptamine type 3 receptors. We used a fluorescence-quenching assay and isothermal titration calorimetry to record low-micromolar dissociation constants for N-methylbicuculline interacting with acetylcholine-binding protein and an engineered version called glycine-binding protein (GBP), which provides a surrogate for the heteromeric interface of the extracellular domain of the glycine receptor (GlyR). The 2.4 Å resolution crystal structure of the GBP:N-methylbicuculline complex, sequence and structural alignments reveal similarities and differences between GlyR and the GABA A receptor-bicuculline interactions. N-methylbicuculline displays a similar conformation in different structures, but adopts distinct orientations enforced by interactions and steric blocks with key residues and plasticity in the binding sites. These features explain the promiscuous activity of bicuculline against the principal inhibitory pentameric ligand-gated ion channels in the CNS.
Organizational Affiliation:
Division of Biological Chemistry and Drug Discovery School of Life Sciences, University of Dundee, Dow St, Dundee, DD1 5EH, UK.