Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Bellini, D., Horrell, S., Hutchin, A., Phippen, C.W., Strange, R.W., Cai, Y., Wagner, A., Webb, J.S., Tews, I., Walsh, M.A.(2017) Sci Rep 7: 42166-42166
- PubMed: 28186120
- DOI: https://doi.org/10.1038/srep42166
- Primary Citation of Related Structures:
4Y9M, 5M1T, 5MF5, 5MFU, 5MKG - PubMed Abstract:
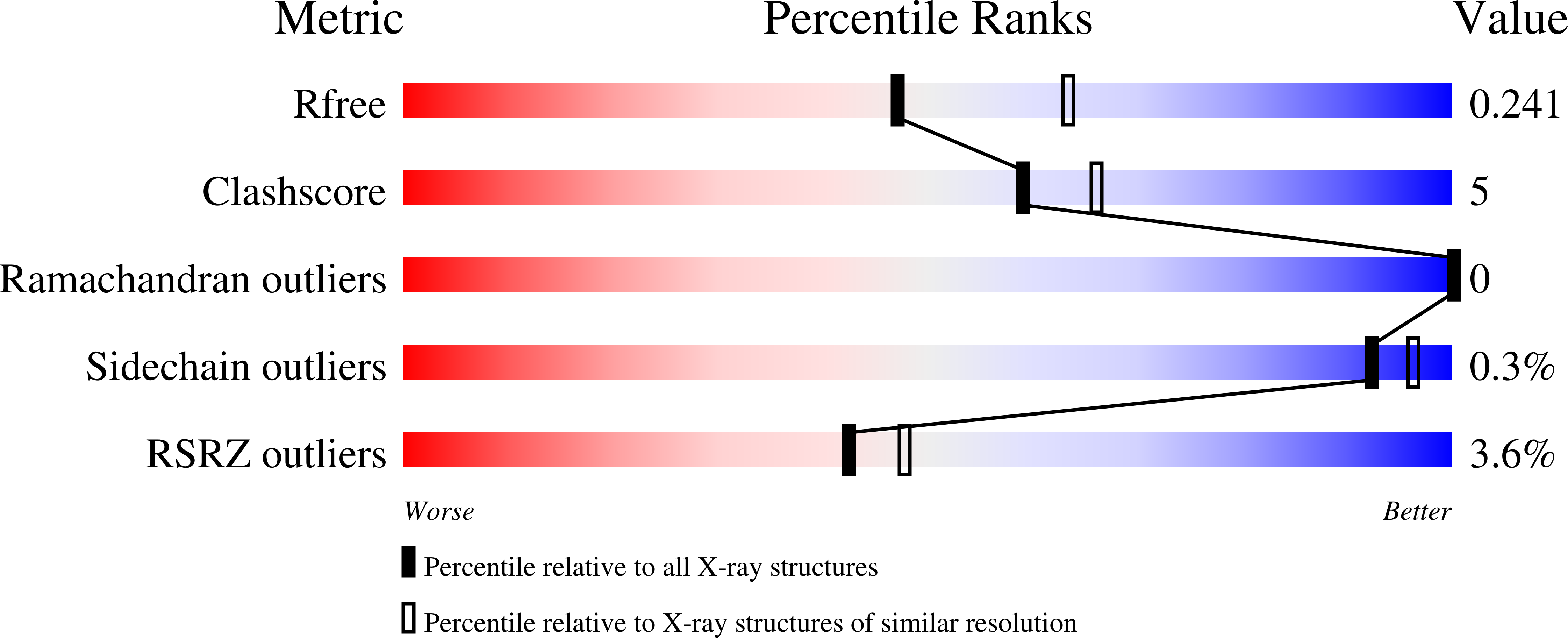
The bacterial second messenger cyclic di-3',5'-guanosine monophosphate (c-di-GMP) is a key regulator of bacterial motility and virulence. As high levels of c-di-GMP are associated with the biofilm lifestyle, c-di-GMP hydrolysing phosphodiesterases (PDEs) have been identified as key targets to aid development of novel strategies to treat chronic infection by exploiting biofilm dispersal. We have studied the EAL signature motif-containing phosphodiesterase domains from the Pseudomonas aeruginosa proteins PA3825 (PA3825 EAL ) and PA1727 (MucR EAL ). Different dimerisation interfaces allow us to identify interface independent principles of enzyme regulation. Unlike previously characterised two-metal binding EAL-phosphodiesterases, PA3825 EAL in complex with pGpG provides a model for a third metal site. The third metal is positioned to stabilise the negative charge of the 5'-phosphate, and thus three metals could be required for catalysis in analogy to other nucleases. This newly uncovered variation in metal coordination may provide a further level of bacterial PDE regulation.
Organizational Affiliation:
Diamond Light Source, Harwell Science and Innovation Campus, Didcot, Oxfordshire, OX11 0FA, United Kingdom.