Structure-guided development of YEATS domain inhibitors by targeting pi-pi-pi stacking.
Li, X., Li, X.M., Jiang, Y., Liu, Z., Cui, Y., Fung, K.Y., van der Beelen, S.H.E., Tian, G., Wan, L., Shi, X., Allis, C.D., Li, H., Li, Y., Li, X.D.(2018) Nat Chem Biol 14: 1140-1149
- PubMed: 30374167
- DOI: https://doi.org/10.1038/s41589-018-0144-y
- Primary Citation of Related Structures:
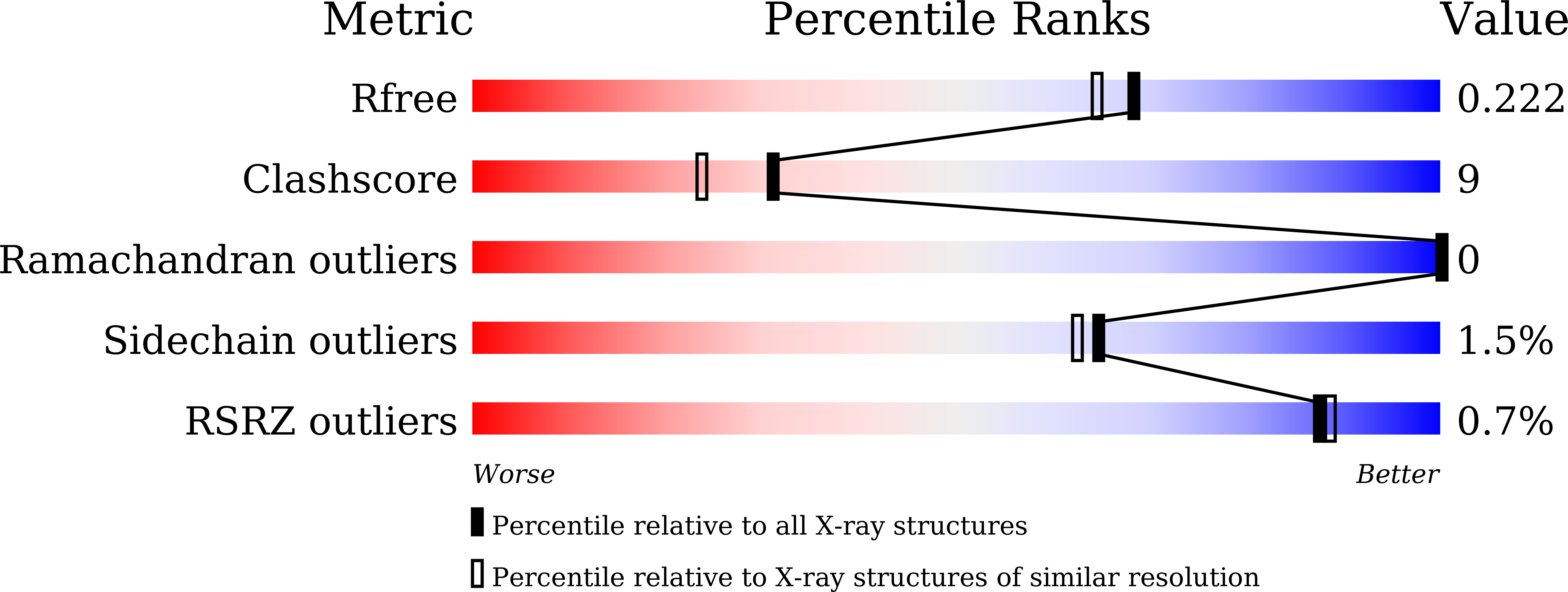
5YYF - PubMed Abstract:
Chemical probes of epigenetic 'readers' of histone post-translational modifications (PTMs) have become powerful tools for mechanistic and functional studies of their target proteins in normal physiology and disease pathogenesis. Here we report the development of the first class of chemical probes of YEATS domains, newly identified 'readers' of histone lysine acetylation (Kac) and crotonylation (Kcr). Guided by the structural analysis of a YEATS-Kcr complex, we developed a series of peptide-based inhibitors of YEATS domains by targeting a unique π-π-π stacking interaction at the proteins' Kcr recognition site. Further structure optimization resulted in the selective inhibitors preferentially binding to individual YEATS-containing proteins including AF9 and ENL with submicromolar affinities. We demonstrate that one of the ENL YEATS-selective inhibitors, XL-13m, engages with endogenous ENL, perturbs the recruitment of ENL onto chromatin, and synergizes the BET and DOT1L inhibition-induced downregulation of oncogenes in MLL-rearranged acute leukemia.
Organizational Affiliation:
Department of Chemistry, The University of Hong Kong, Hong Kong, China.