Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin.
Bowden, C.F.M., Chan, A.C.K., Li, E.J.W., Arrieta, A.L., Eltis, L.D., Murphy, M.E.P.(2018) J Biol Chem 293: 177-190
- PubMed: 29109153
- DOI: https://doi.org/10.1074/jbc.M117.806562
- Primary Citation of Related Structures:
5VMM - PubMed Abstract:
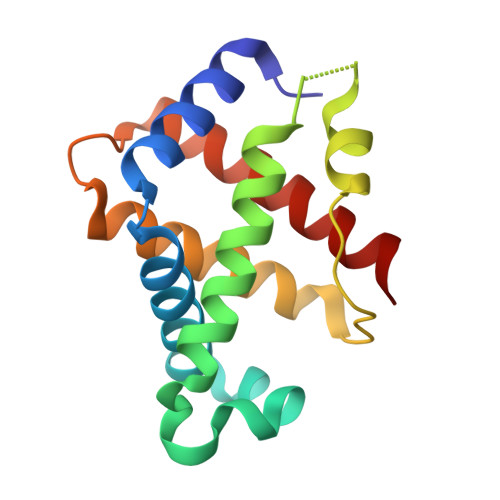
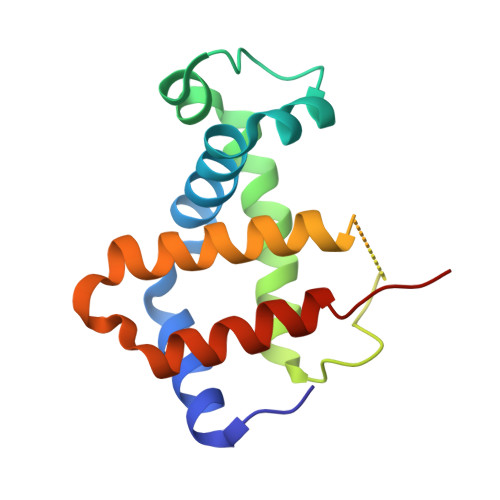
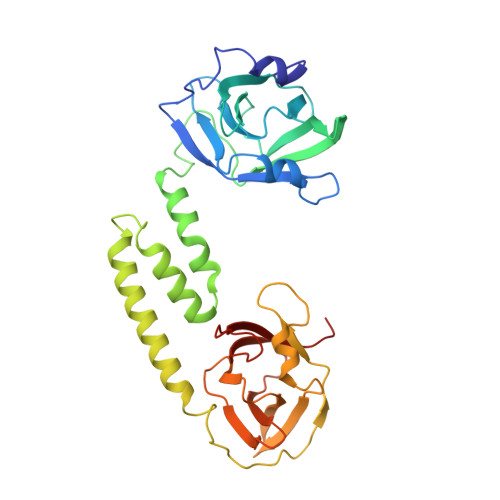
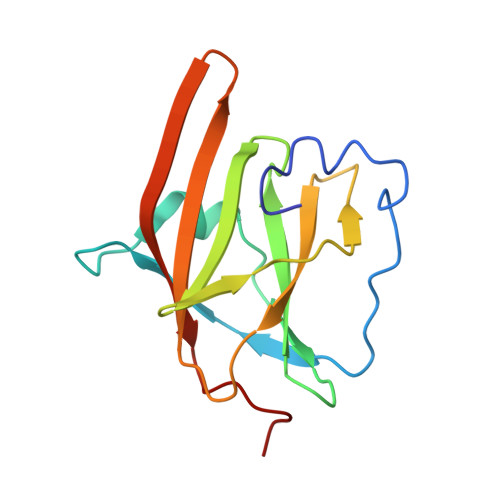
IsdB is a receptor on the surface of the bacterial pathogen Staphylococcus aureus that extracts heme from hemoglobin (Hb) to enable growth on Hb as a sole iron source. IsdB is critically important both for in vitro growth on Hb and in infection models and is also highly up-regulated in blood, serum, and tissue infection models, indicating a key role of this receptor in bacterial virulence. However, structural information for IsdB is limited. We present here a crystal structure of a complex between human Hb and IsdB. In this complex, the α subunits of Hb are refolded with the heme displaced to the interface with IsdB. We also observe that atypical residues of Hb, His 58 and His 89 of αHb, coordinate to the heme iron, which is poised for transfer into the heme-binding pocket of IsdB. Moreover, the porphyrin ring interacts with IsdB residues Tyr 440 and Tyr 444 Previously, Tyr 440 was observed to coordinate heme iron in an IsdB·heme complex structure. A Y440F/Y444F IsdB variant we produced was defective in heme transfer yet formed a stable complex with Hb ( K d = 6 ± 2 μm) in solution with spectroscopic features of the bis-His species observed in the crystal structure. Haptoglobin binds to a distinct site on Hb to inhibit heme transfer to IsdB and growth of S. aureus , and a ternary complex of IsdB·Hb·Hp was observed. We propose a model for IsdB heme transfer from Hb that involves unfolding of Hb and heme iron ligand exchange.
Organizational Affiliation:
Department of Microbiology and Immunology, Life Sciences Institute, University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada.