Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
De Santi, C., Leiros, H.S., Di Scala, A., De Pascale, D., Altermark, B., Willassen, N.(2016) Extremophiles 20: 323
- PubMed: 27016194
- DOI: https://doi.org/10.1007/s00792-016-0824-z
- Primary Citation of Related Structures:
4V2I - PubMed Abstract:
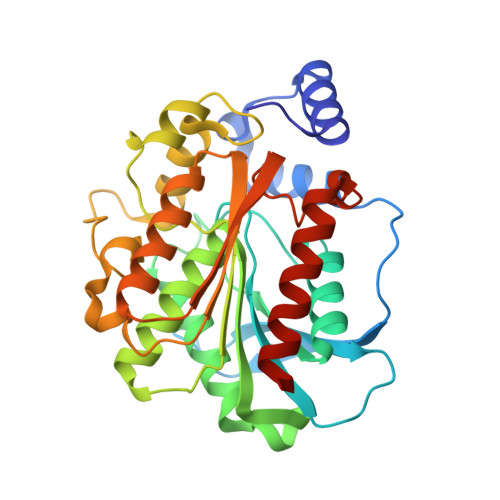
A gene encoding an esterase, ThaEst2349, was identified in the marine psychrophilic bacterium Thalassospira sp. GB04J01. The gene was cloned and overexpressed in E. coli as a His-tagged fusion protein. The recombinant enzyme showed optimal activity at 45 °C and the thermal stability displayed a retention of 75 % relative activity at 40 °C after 2 h. The optimal pH was 8.5 but the enzyme kept more than 75 % of its maximal activity between pH 8.0 and 9.5. ThaEst2349 also showed remarkable tolerance towards high concentrations of salt and it was active against short-chain p-nitrophenyl esters, displaying optimal activity with the acetate. The enzyme was tested for tolerance of organic solvents and the results are suggesting that it could function as an interesting candidate for biotechnological applications. The crystal structure of ThaEst2349 was determined to 1.69 Å revealing an asymmetric unit containing two chains, which also is the biological unit. The structure has a characteristic cap domain and a catalytic triad comprising Ser158, His285 and Asp255. To explain the cold-active nature of the enzyme, we compared it against thermophilic counterparts. Our hypothesis is that a high methionine content, less hydrogen bonds and less ion pairs render the enzyme more flexible at low temperatures.
Organizational Affiliation:
Institute of Protein Biochemistry, National Research Council, Via P. Castellino, 111., 80131, Naples, Italy. concetta.d.santi@uit.no.