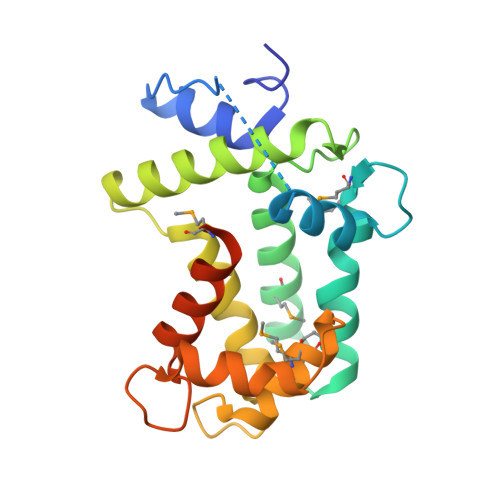
Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Stepanyuk, G.A., Liu, Z.J., Burakova, L.P., Lee, J., Rose, J., Vysotski, E.S., Wang, B.C.(2013) Biochim Biophys Acta 1834: 2139-2146
- PubMed: 23891746
- DOI: https://doi.org/10.1016/j.bbapap.2013.07.006
- Primary Citation of Related Structures:
4MN0 - PubMed Abstract:
The bright bioluminescence of ctenophores, found in oceans worldwide, is determined by Ca(2+)-regulated photoproteins, functionally identical to and sharing many properties of hydromedusan photoproteins. In contrast, however, the ctenophore photoproteins are extremely sensitive to UV and visible light over the range of their absorption spectrum. The spatial structure of a novel light-sensitive photoprotein from the ctenophore Beroe abyssicola in its apoform bound with three calcium ions is determined at 2.0Å. We demonstrate that the apoberovin is a slightly asymmetrical compact globular protein formed by two domains with a cavity in the center, which exactly retains the fold architecture characteristic of hydromedusan photoproteins despite their low amino acid sequence identity. However, the structural alignment of these two photoprotein classes clearly shows that despite the high similarity of shape and geometry of their coelenterazine-binding cavities, their interiors differ drastically. The key residues appearing to be crucial for stabilizing the 2-hydroperoxycoelenterazine and for formation of the emitter in hydromedusan photoproteins, are replaced in berovin by amino acid residues having completely different side chain properties. Evidently, these replacements must be responsible for the distinct properties of ctenophore photoproteins such as sensitivity to light or the fact that the formation of active photoprotein from apophotoprotein, coelenterazine, and oxygen is more effective at alkaline pH.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602, USA; Photobiology Laboratory, Institute of Biophysics Russian Academy of Sciences, Siberian Branch, Krasnoyarsk 660036, Russia.