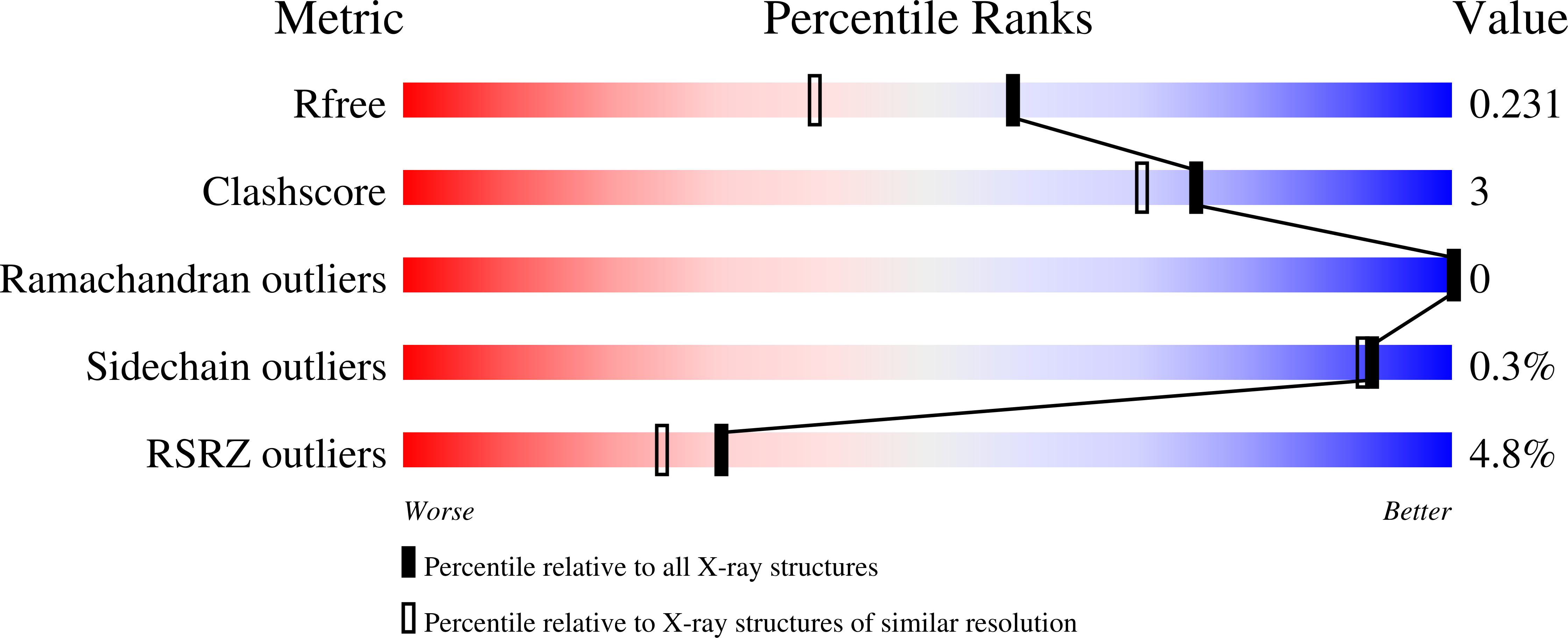
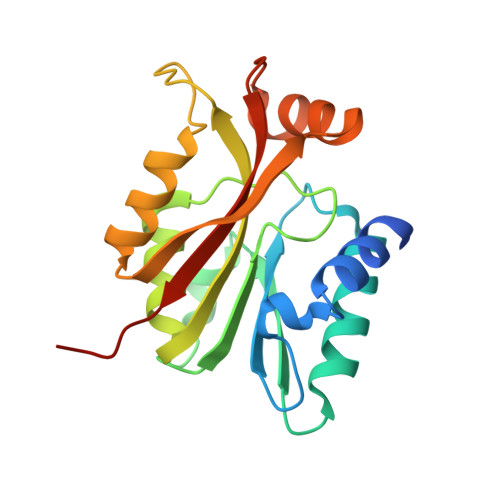
Crystal structure of the N-terminal methyltransferase-like domain of anamorsin.
Song, G., Cheng, C., Li, Y., Shaw, N., Xiao, Z.C., Liu, Z.J.(2014) Proteins 82: 1066-1071
- PubMed: 24123282
- DOI: https://doi.org/10.1002/prot.24443
- Primary Citation of Related Structures:
4M7R - PubMed Abstract:
Anamorsin is a recently identified molecule that inhibits apoptosis during hematopoiesis. It contains an N-terminal methyltransferase-like domain and a C-terminal Fe-S cluster motif. Not much is known about the function of the protein. To better understand the function of anamorsin, we have solved the crystal structure of the N-terminal domain at 1.8 Å resolution. Although the overall structure resembles a typical S-adenosylmethionine (SAM) dependent methyltransferase fold, it lacks one α-helix and one β-strand. As a result, the N-terminal domain as well as the full-length anamorsin did not show S-adenosyl-L-methionine (AdoMet) dependent methyltransferase activity. Structural comparisons with known AdoMet dependent methyltransferases reveals subtle differences in the SAM binding pocket that preclude the N-terminal domain from binding to AdoMet. The N-terminal methyltransferase-like domain of anamorsin probably functions as a structural scaffold to inhibit methyl transfers by out-competing other AdoMet dependant methyltransferases or acts as bait for protein-protein interactions.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.