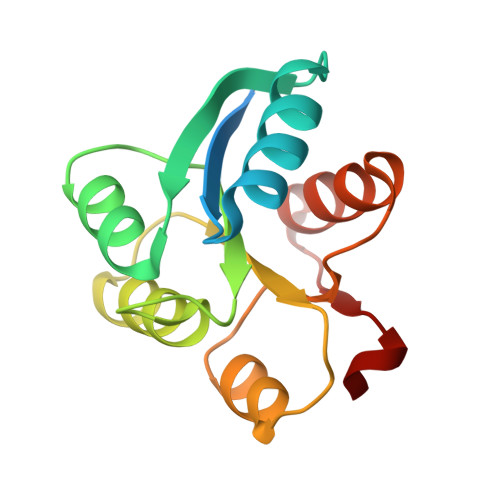
Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the absence and presence of the phosphoryl analog beryllofluoride.
Park, A.K., Moon, J.H., Lee, K.S., Chi, Y.M.(2012) Biochem Biophys Res Commun 421: 403-407
- PubMed: 22521891
- DOI: https://doi.org/10.1016/j.bbrc.2012.04.035
- Primary Citation of Related Structures:
4E7O, 4E7P - PubMed Abstract:
Spr1814 of Streptococcus pneumoniae is a putative response regulator (RR) that has four-helix helix-turn-helix DNA-binding domain and belongs to the NarL family. The prototypical RR contains two domains, an N-terminal receiver domain linked to a variable effector domain. The receiver domain functions as a phosphorylation-activated switch and contains the typical doubly wound five-stranded α/β fold. Here, we report the crystal structure of the receiver domain of spr1814 (spr1814(R)) determined in the absence and presence of beryllofluoride as a phosphoryl analog. Based on the overall structure, spr1814(R) was shown to contain the typical fold similar with other structures of the receiver domain; however, an additional linker region connecting the receiver and DNA-binding domain was inserted into the dimer interface of spr1814(R), resulting in the formation of unique dimer interface. Upon phosphorylation, the conformational change of the linker region was observed and this suggests that domain rearrangement between the receiver domain and effector domain could occur in full-length spr1814.
Organizational Affiliation:
Division of Biotechnology, College of Life Sciences, Korea University, Seoul 136-713, Republic of Korea.