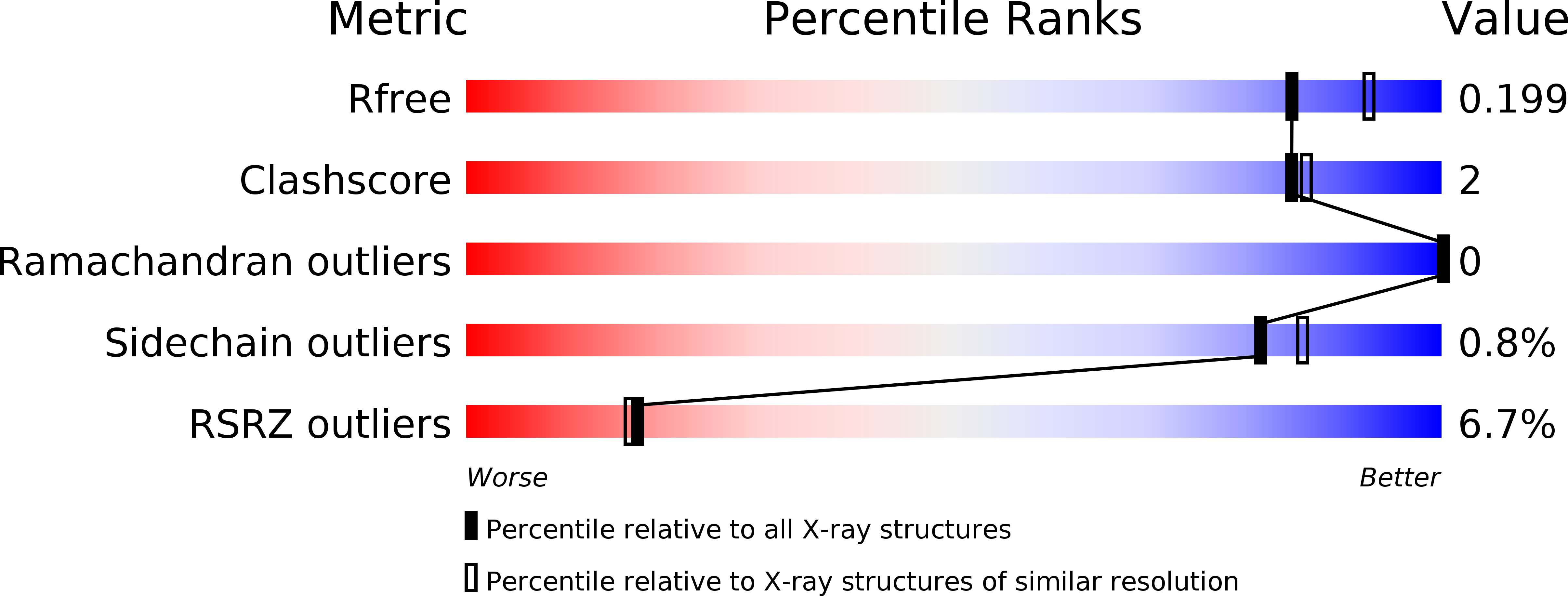
Entry History & Funding Information
Deposition Data
- Released Date: 2015-05-27
Deposition Author(s): Michalska, K., Bigelow, L., Jedrzejczak, R., Babnigg, G., Lohman, J., Ma, M., Rudolf, J., Chang, C.-Y., Shen, B., Joachimiak, A., Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro)
| Funding Organization | Location | Grant Number |
|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM094585 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM098248 |
- Version 1.0: 2015-05-27
Type: Initial release
- Version 1.1: 2016-09-21
Changes: Source and taxonomy - Version 1.2: 2017-09-06
Changes: Author supporting evidence, Derived calculations - Version 1.3: 2017-11-22
Changes: Refinement description - Version 1.4: 2019-12-25
Changes: Author supporting evidence