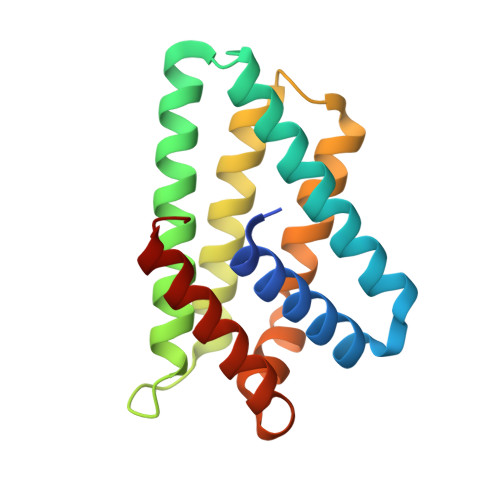
Crystal structure of TIPE2 provides insights into immune homeostasis
Zhang, X., Wang, J., Fan, C., Li, H., Sun, H., Gong, S., Chen, Y.H., Shi, Y.(2009) Nat Struct Mol Biol 16: 89-90
- PubMed: 19079267
- DOI: https://doi.org/10.1038/nsmb.1522
- Primary Citation of Related Structures:
3F4M - PubMed Abstract:
TNFAIP8-like 2 (TIPE2) has an essential role in immune homeostasis, yet the underlying mechanism remains enigmatic. The high-resolution crystal structure of TIPE2 reveals a previously uncharacterized fold that is different from the predicted fold of a death effector domain (DED). Strikingly, TIPE2 contains a large, hydrophobic central cavity that is poised for cofactor binding. These structural features will be important for understanding the functions of TIPE2 and other TNFAIP8 family proteins.
Organizational Affiliation:
Center for Structural Biology and Department of Biological Sciences and Biotechnology, Tsinghua University, Beijing 100084, China.