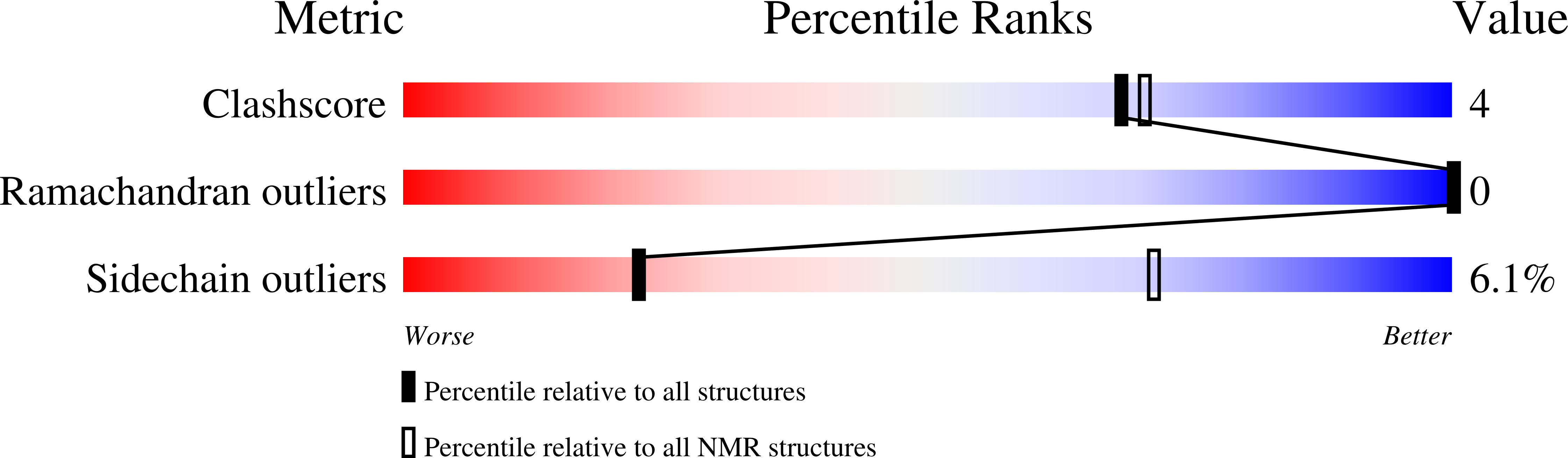Nucleosomal DNA Binding Drives the Recognition of H3K36-Methylated Nucleosomes by the Psip1-Pwwp Domain.
Van Nuland, R., Van Schaik, F.M., Simonis, M., Van Heesch, S., Cuppen, E., Boelens, R., Timmers, H.T.M., Van Ingen, H.(2013) Epigenetics Chromatin 6: 12
- PubMed: 23656834
- DOI: https://doi.org/10.1186/1756-8935-6-12
- Primary Citation of Related Structures:
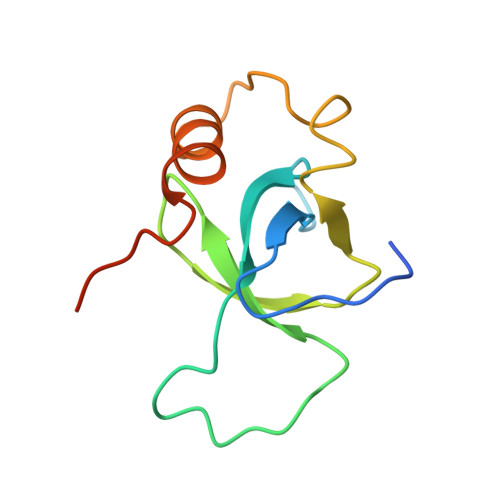
3ZEH - PubMed Abstract:
Recognition of histone modifications by specialized protein domains is a key step in the regulation of DNA-mediated processes like gene transcription. The structural basis of these interactions is usually studied using histone peptide models, neglecting the nucleosomal context. Here, we provide the structural and thermodynamic basis for the recognition of H3K36-methylated (H3K36me) nucleosomes by the PSIP1-PWWP domain, based on extensive mutational analysis, advanced nuclear magnetic resonance (NMR), and computational approaches.
Organizational Affiliation:
NMR Spectroscopy Research Group, Bijvoet Center for Biomolecular Research, Utrecht University Utrecht, Padualaan 8, Utrecht, CH, 3854, The Netherlands. h.vaningen@uu.nl.