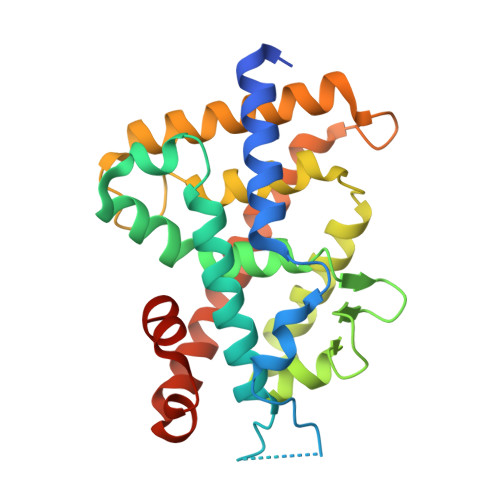
Design, synthesis and X-ray crystallographic study of new nonsecosteroidal vitamin D receptor ligands
Demizu, Y., Takahashi, T., Kaneko, F., Sato, Y., Okuda, H., Ochiai, E., Horie, K., Takagi, K., Kakuda, S., Takimoto-Kamimura, M., Kurihara, M.(2011) Bioorg Med Chem Lett 21: 6104-6107
- PubMed: 21889334
- DOI: https://doi.org/10.1016/j.bmcl.2011.08.047
- Primary Citation of Related Structures:
3AUN - PubMed Abstract:
We designed and synthesized nonsecosteroidal vitamin D receptor (VDR) ligands that formed H-bonds with six amino acid residues (Tyr143, Ser233, Arg270, Ser274, His301 and His393) of the VDR ligand-binding domain. The ligand YR335 exhibited potent transcriptional activity, which was comparable to those of 1α,25-dihydroxyvitamin D(3) and YR301. The crystal structure of the complex formed between YR335 and the VDR ligand-binding domain was solved, which revealed that YR335 formed H-bonds with the six amino acid residues mentioned above.
Organizational Affiliation:
Division of Organic Chemistry, National Institute of Health Sciences, 1-18-1, Kamiyoga, Setagaya, Tokyo 158-8501, Japan.