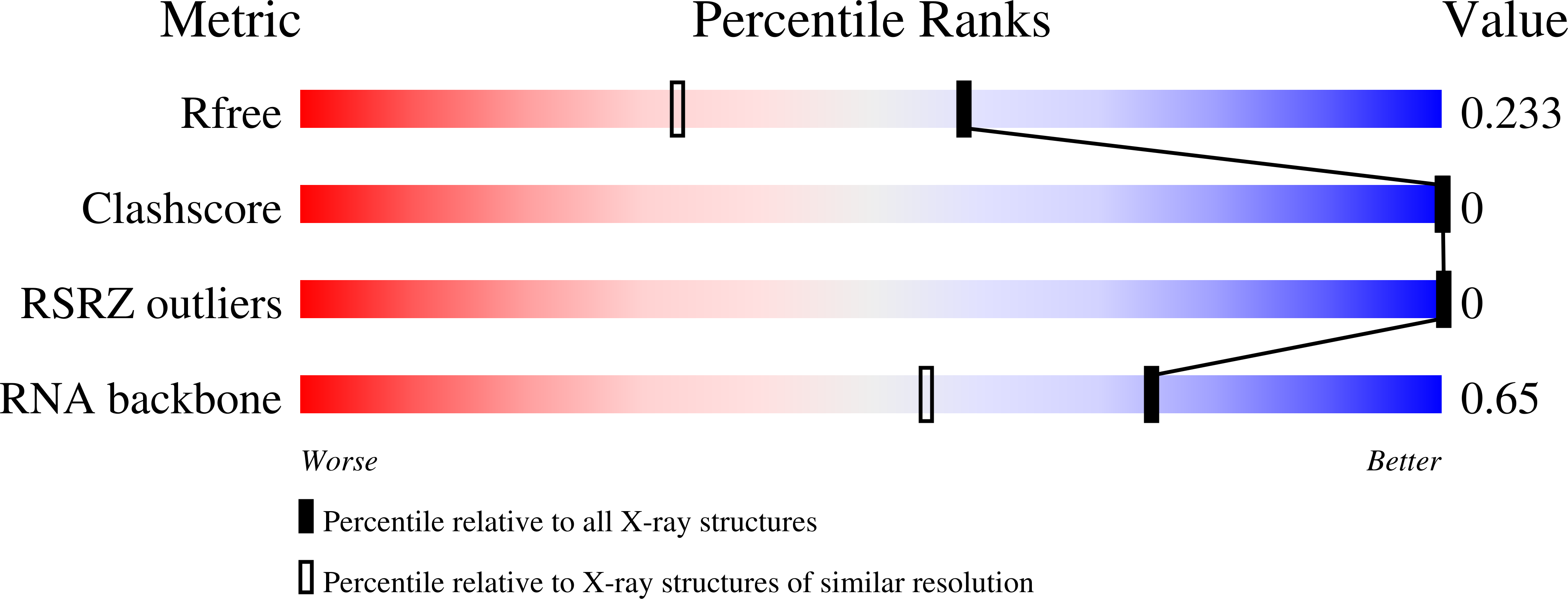Deduced RNA binding mechanism of ThiI based on structural and binding analyses of a minimal RNA ligand
Tanaka, Y., Yamagata, S., Kitago, Y., Yamada, Y., Chimnaronk, S., Yao, M., Tanaka, I.(2009) RNA 15: 1498-1506
- PubMed: 19509301
- DOI: https://doi.org/10.1261/rna.1614709
- Primary Citation of Related Structures:
2ZY6 - PubMed Abstract:
ThiI catalyzes the thio-introduction reaction to tRNA, and a truncated tRNA consisting of 39 nucleotides, TPHE39A, is the minimal RNA substrate for modification by ThiI from Escherichia coli. To examine the molecular basis of the tRNA recognition by ThiI, we have solved the crystal structure of TPHE39A, which showed that base pairs in the T-stem were almost completely disrupted, although those in the acceptor-stem were preserved. Gel shift assays and isothermal titration calorimetry experiments showed that ThiI can efficiently bind with not only tRNA(Phe) but also TPHE39A. Binding assays using truncated ThiI, i.e., N- and C-terminal domains of ThiI, showed that the N-domain can bind with both tRNA(Phe) and TPHE39A, whereas the C-domain cannot. These results indicated that the N-domain of ThiI recognizes the acceptor-stem region. Thermodynamic analysis indicated that the C-domain also affects RNA binding by its enthalpically favorable, but entropically unfavorable, contribution. In addition, circular dichroism spectra showed that the C-domain induced a conformation change in tRNA(Phe). Based on these results, a possible RNA binding mechanism of ThiI in which the N-terminal domain recognizes the acceptor-stem region and the C-terminal region causes a conformational change of RNA is proposed.
Organizational Affiliation:
Creative Research Institute Sousei, Hokkaido University, Sapporo, 001-0021, Japan. tanaka@castor.sci.hokudai.ac.jp