Effects of Salt on the Kinetics and Thermodynamic Stability of Endonuclease I from Vibrio Salmonicida and Vibrio Cholerae.
Niiranen, L., Altermark, B., Brandsdal, B.O., Leiros, H.-K.S., Helland, R., Smalas, A.O., Willassen, N.P.(2008) FEBS J 275: 1593
- PubMed: 18312415
- DOI: https://doi.org/10.1111/j.1742-4658.2008.06317.x
- Primary Citation of Related Structures:
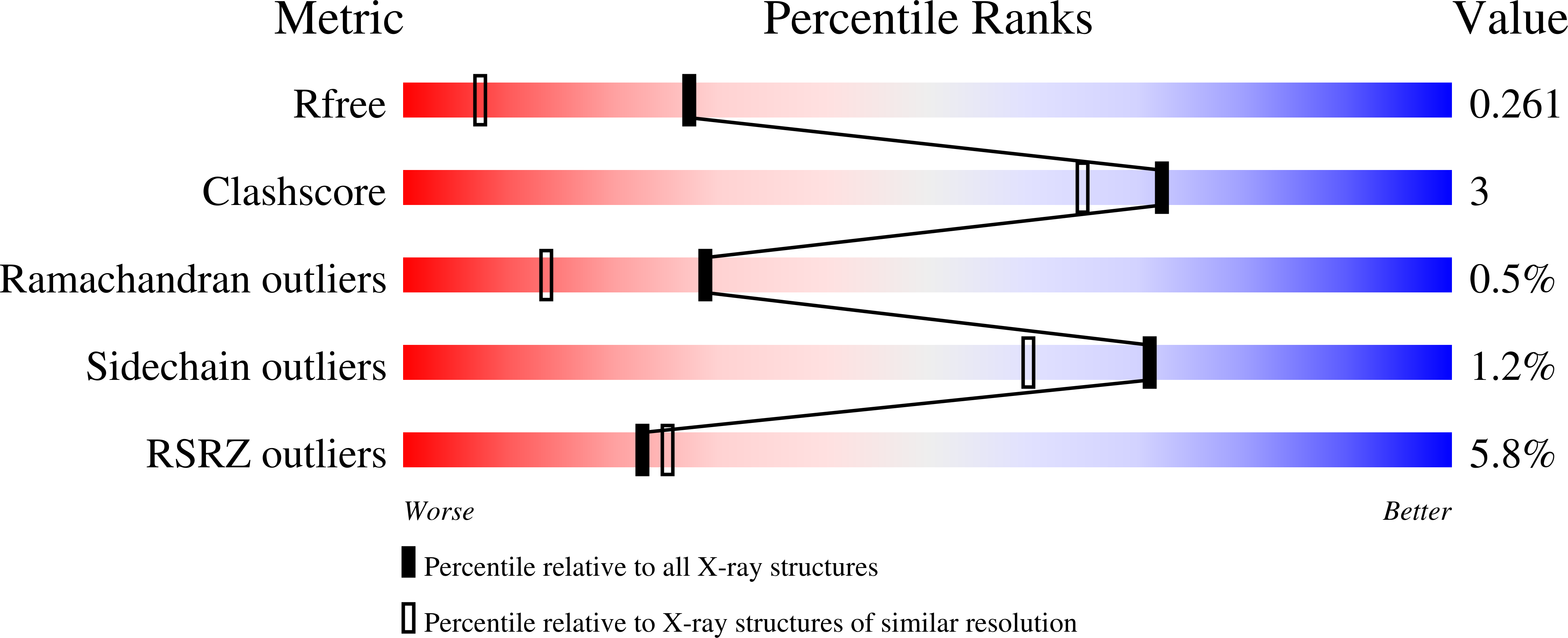
2VND - PubMed Abstract:
Adaptation to extreme environments affects the stability and catalytic efficiency of enzymes, often endowing them with great industrial potential. We compared the environmental adaptation of the secreted endonuclease I from the cold-adapted marine fish pathogen Vibrio salmonicida (VsEndA) and the human pathogen Vibrio cholerae (VcEndA). Kinetic analysis showed that VsEndA displayed unique halotolerance. It retained a considerable amount of activity from low concentrations to at least 0.6 m NaCl, and was adapted to work at higher salt concentrations than VcEndA by maintaining a low K(m) value and increasing k(cat). In differential scanning calorimetry, salt stabilized both enzymes, but the effect on the calorimetric enthalpy and cooperativity of unfolding was larger for VsEndA, indicating salt dependence. Mutation of DNA binding site residues (VsEndA, Q69N and K71N; VcEndA, N69Q and N71K) affected the kinetic parameters. The VsEndA Q69N mutation also increased the T(m) value, whereas other mutations affected mainly DeltaH(cal). The determined crystal structure of VcEndA N69Q revealed the loss of one hydrogen bond present in native VcEndA, but also the formation of a new hydrogen bond involving residue 69 that could possibly explain the similar T(m) values for native and N69Q-mutated VcEndA. Structural analysis suggested that the stability, catalytic efficiency and salt tolerance of EndA were controlled by small changes in the hydrogen bonding networks and surface electrostatic potential. Our results indicate that endonuclease I adaptation is closely coupled to the conditions of the habitats of natural Vibrio, with VsEndA displaying a remarkable salt tolerance unique amongst the endonucleases characterized so far.
Organizational Affiliation:
Department of Molecular Biotechnology, Institute of Medical Biology, Faculty of Medicine, University of Tromsø, Norway.