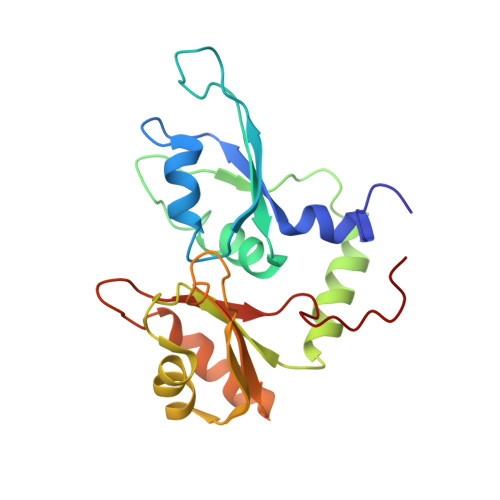
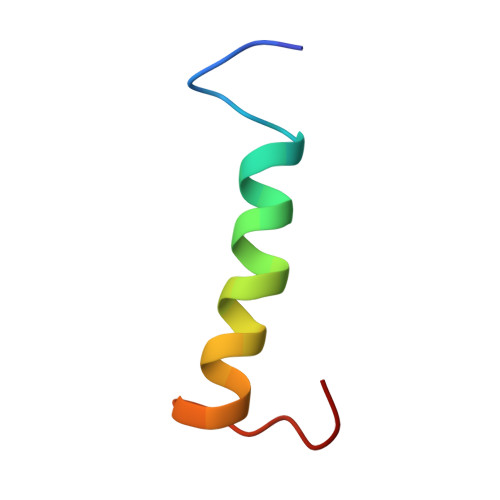
Molecular basis of FIR-mediated c-myc transcriptional control.
Cukier, C.D., Hollingworth, D., Martin, S.R., Kelly, G., Diaz-Moreno, I., Ramos, A.(2010) Nat Struct Mol Biol 17: 1058-1064
- PubMed: 20711187
- DOI: https://doi.org/10.1038/nsmb.1883
- Primary Citation of Related Structures:
2KXF, 2KXH - PubMed Abstract:
The far upstream element (FUSE) regulatory system promotes a peak in the concentration of c-Myc during cell cycle. First, the FBP transcriptional activator binds to the FUSE DNA element upstream of the c-myc promoter. Then, FBP recruits its specific repressor (FIR), which acts as an on/off transcriptional switch. Here we describe the molecular basis of FIR recruitment, showing that the tandem RNA recognition motifs of FIR provide a platform for independent FUSE DNA and FBP protein binding and explaining the structural basis of the reversibility of the FBP-FIR interaction. We also show that the physical coupling between FBP and FIR is modulated by a flexible linker positioned sequentially to the recruiting element. Our data explain how the FUSE system precisely regulates c-myc transcription and suggest that a small change in FBP-FIR affinity leads to a substantial effect on c-Myc concentration.
Organizational Affiliation:
Molecular Structure Division, Medical Research Council National Institute for Medical Research, London, UK.