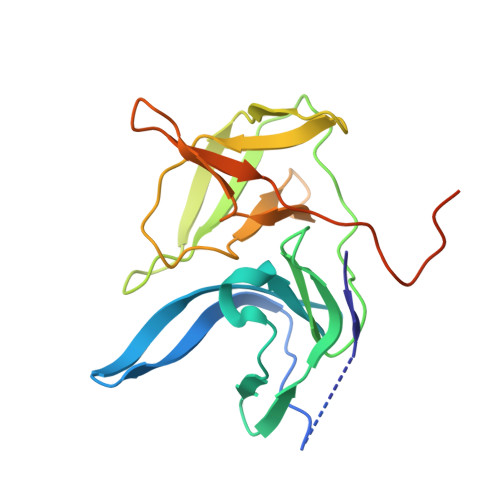
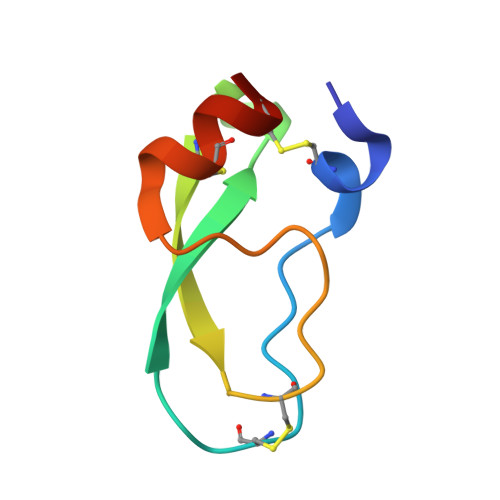
Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold.
Aleshin, A.E., Shiryaev, S.A., Strongin, A.Y., Liddington, R.C.(2007) Protein Sci 16: 795-806
- PubMed: 17400917
- DOI: https://doi.org/10.1110/ps.072753207
- Primary Citation of Related Structures:
2GGV, 2IJO - PubMed Abstract:
Pathogenic members of the flavivirus family, including West Nile Virus (WNV) and Dengue Virus (DV), are growing global threats for which there are no specific treatments. The two-component flaviviral enzyme NS2B-NS3 cleaves the viral polyprotein precursor within the host cell, a process that is required for viral replication. Here, we report the crystal structure of WNV NS2B-NS3pro both in a substrate-free form and in complex with the trypsin inhibitor aprotinin/BPTI. We show that aprotinin binds in a substrate-mimetic fashion in which the productive conformation of the protease is fully formed, providing evidence for an "induced fit" mechanism of catalysis and allowing us to rationalize the distinct substrate specificities of WNV and DV proteases. We also show that the NS2B cofactor of WNV can adopt two very distinct conformations and that this is likely to be a general feature of flaviviral proteases, providing further opportunities for regulation. Finally, by comparing the flaviviral proteases with the more distantly related Hepatitis C virus, we provide insights into the evolution of the Flaviviridae fold. Our work should expedite the design of protease inhibitors to treat a range of flaviviral infections.
Organizational Affiliation:
Infectious and Inflammatory Disease Center, Burnham Institute for Medical Research, La Jolla, California 92037, USA.