Insight Into S-Adenosylmethionine Biosynthesis from the Crystal Structures of the Human Methionine Adenosyltransferase Catalytic and Regulatory Subunits.
Shafqat, N., Muniz, J.R.C., Pilka, E.S., Papagrigoriou, E., von Delft, F., Oppermann, U., Yue, W.W.(2013) Biochem J 452: 27
- PubMed: 23425511
- DOI: https://doi.org/10.1042/BJ20121580
- Primary Citation of Related Structures:
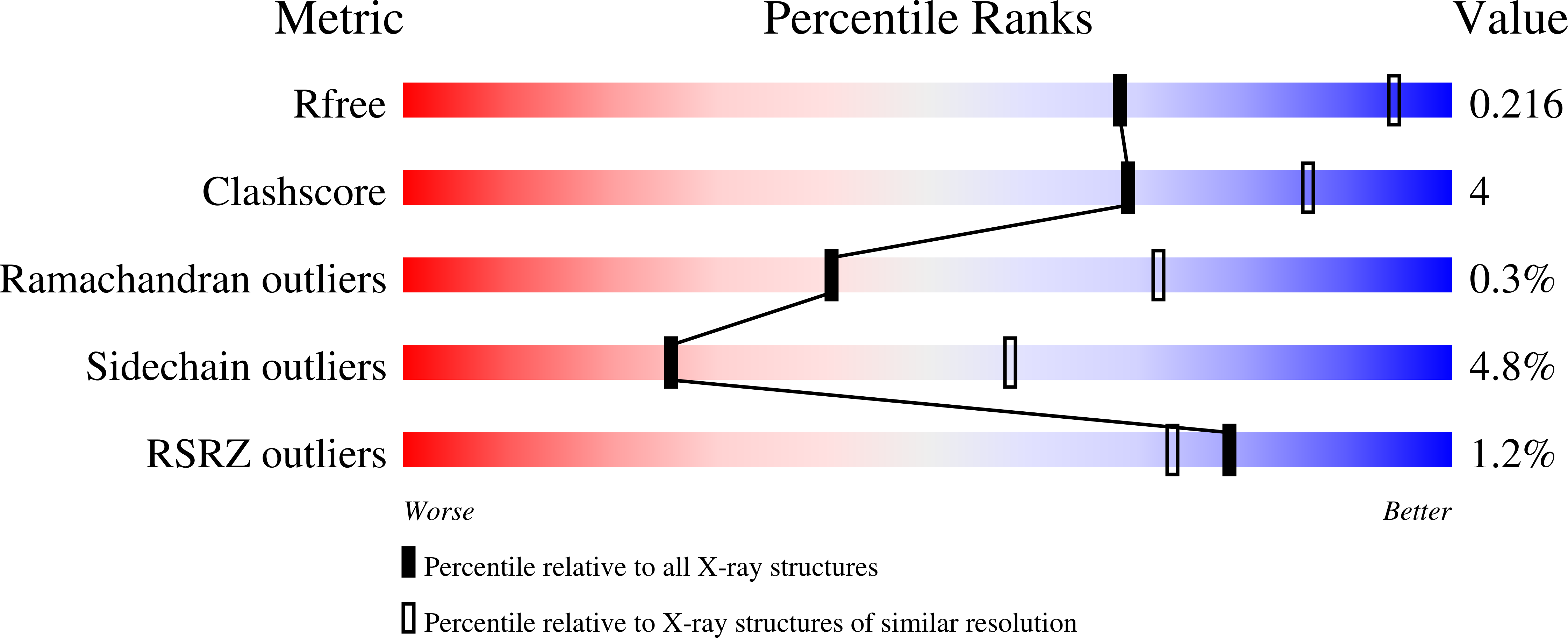
2YDX, 2YDY - PubMed Abstract:
MAT (methionine adenosyltransferase) utilizes L-methionine and ATP to form SAM (S-adenosylmethionine), the principal methyl donor in biological methylation. Mammals encode a liver-specific isoenzyme, MAT1A, that is genetically linked with an inborn metabolic disorder of hypermethioninaemia, as well as a ubiquitously expressed isoenzyme, MAT2A, whose enzymatic activity is regulated by an associated subunit MAT2B. To understand the molecular mechanism of MAT functions and interactions, we have crystallized the ligand-bound complexes of human MAT1A, MAT2A and MAT2B. The structures of MAT1A and MAT2A in binary complexes with their product SAM allow for a comparison with the Escherichia coli and rat structures. This facilitates the understanding of the different substrate or product conformations, mediated by the neighbouring gating loop, which can be accommodated by the compact active site during catalysis. The structure of MAT2B reveals an SDR (short-chain dehydrogenase/reductase) core with specificity for the NADP/H cofactor, and harbours the SDR catalytic triad (YxxxKS). Extended from the MAT2B core is a second domain with homology with an SDR sub-family that binds nucleotide-sugar substrates, although the equivalent region in MAT2B presents a more open and extended surface which may endow a different ligand/protein-binding capability. Together, the results of the present study provide a framework to assign structural features to the functional and catalytic properties of the human MAT proteins, and facilitate future studies to probe new catalytic and binding functions.
Organizational Affiliation:
Structural Genomics Consortium, University of Oxford, Oxford OX3 7DQ, UK.