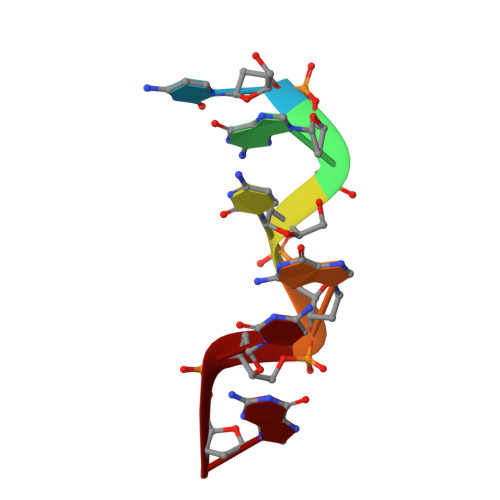Interaction between the left-handed Z-DNA and polyamine-2. The crystal structure of the d(CG)3 and spermidine complex.
Ohishi, H., Nakanishi, I., Inubushi, K., van der Marel, G., van Boom, J.H., Rich, A., Wang, A.H., Hakoshima, T., Tomita, K.(1996) FEBS Lett 391: 153-156
- PubMed: 8706905
- DOI: https://doi.org/10.1016/0014-5793(96)00723-5
- Primary Citation of Related Structures:
293D - PubMed Abstract:
This paper deals with the crystal structure of d(CG)3-spermidine complex. The DNA fragment, d(CG)3, was crystallized with N-(2-amino-propyl)-1,4-diamino-butane, PA(34), spermidine. The results of its X-ray crystallographic analysis showed many intermolecular contacts between d(CG)3 and spermidine, but the binding mode of spermidine to the d(CG)3 molecule is different from that of the d(CG)3 and N-(2-amino-ethyl)-1,4-diamino-butane [PA(24)] complex: a spermidine molecule bound to the d(CG)3 and its symmetrically related neighboring d(CG)3 molecules through the water molecules with hydrogen bonds, while one PA(24) molecule connected directly to one d(CG)3 molecule, but not to its neighboring d(CG)3 molecule. In the crystal, the d(CG)3 molecule was the left-handed Z-form, and three magnesium cations and a sodium cation were observed around the d(CG)3 moiety with different binding modes from the case of the d(CG)3-PA(24) complex.
Organizational Affiliation:
Osaka University of Pharmaceutical Sciences, Japan. ohishi@oysun01.oups.ac.jp