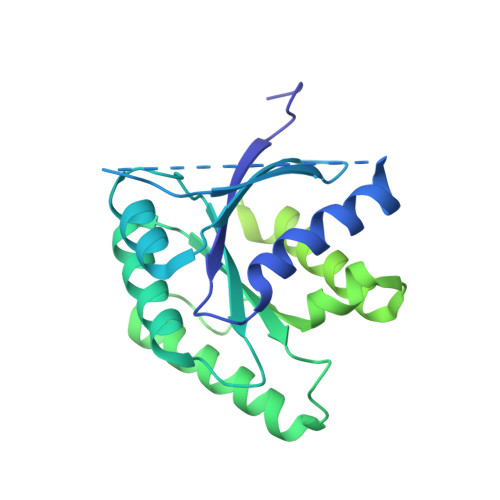
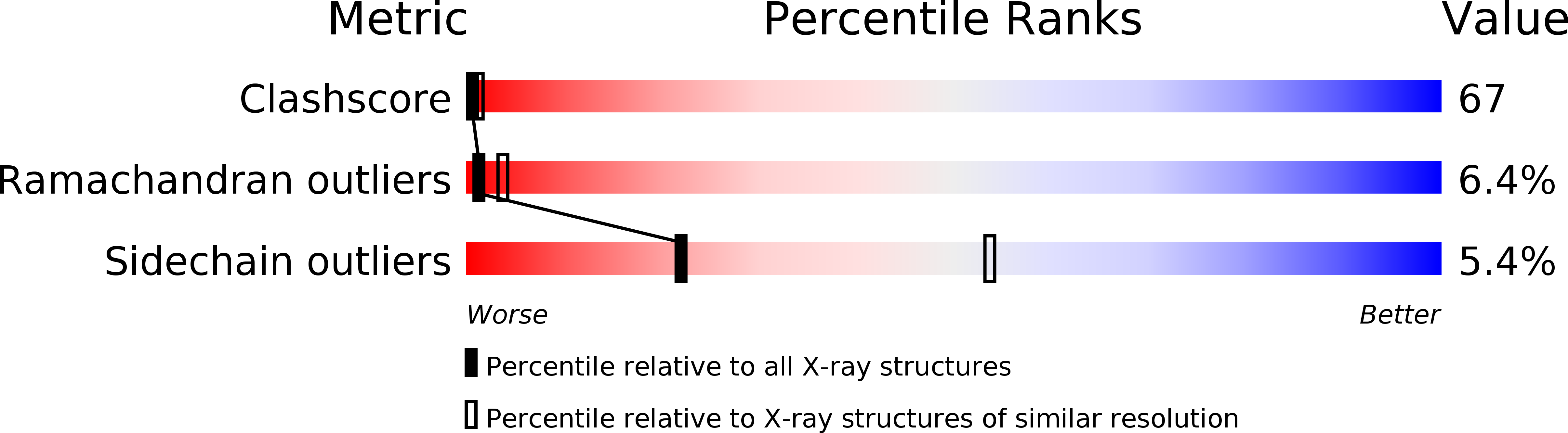Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography.
la Cour, T.F., Nyborg, J., Thirup, S., Clark, B.F.(1985) EMBO J 4: 2385-2388
- PubMed: 3908095
- DOI: https://doi.org/10.1002/j.1460-2075.1985.tb03943.x
- Primary Citation of Related Structures:
1ETU - PubMed Abstract:
Structural details of the guanosine diphosphate binding to a modified form of elongation factor Tu from Escherichia coli, resulting from X-ray crystallographic studies, are reported. The protein elements that take part in the nucleotide binding are located in four loops connecting beta-strands with alpha-helices. These loops correspond to regions in primary sequences which show a high degree of homology when compared with other prokaryotic and eukaryotic elongation factors and initiation factor 2.