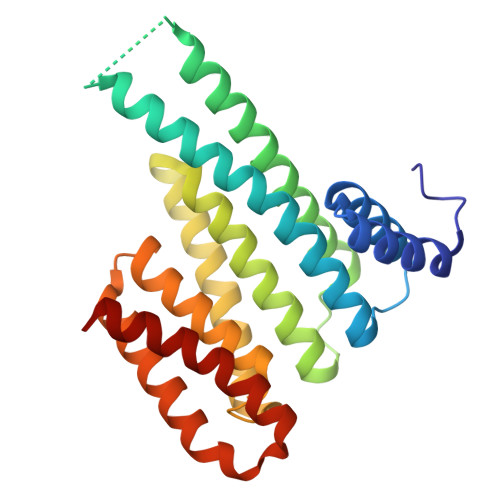
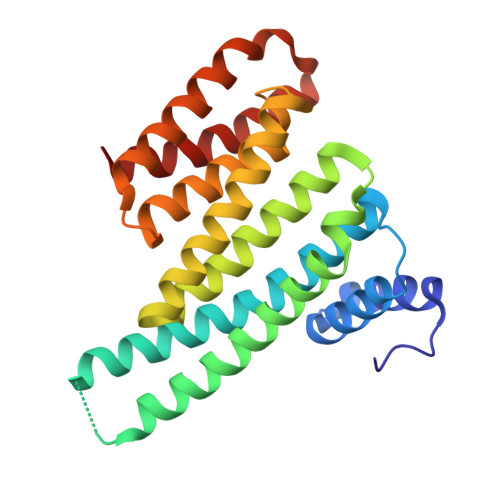
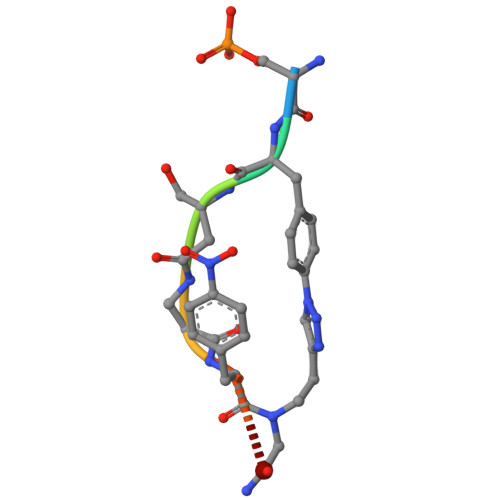
Macrocyclic Molecular Glues for the 14-3-3/ChREBP Interaction: Affinity and Cooperativity in an Inverse Relationship.
Pennings, M.A.M., van den Bosch, M.A.W., Oberheide, A., Verhoef, C.J.A., Ottmann, C., Markvoort, A.J., Miley, G.P., Brunsveld, L.(2025) Angew Chem Int Ed Engl : e21678-e21678
- PubMed: 41414041
- DOI: https://doi.org/10.1002/anie.202521678
- Primary Citation of Related Structures:
9SA9, 9SAB, 9SAE - PubMed Abstract:
Molecular glues (MGs) stabilize protein-protein interactions (PPIs) by simultaneously binding two or more proteins at their composite interface. Macrocycles present attractive properties as MGs, including large contact surfaces to address the often flat and undefined composite PPI interfaces, but their structure-based design has remained intangible. We have designed peptidomimetic macrocycles capable of enhancing the PPI between 14-3-3 and the carbohydrate response element binding protein (ChREBP), a regulatory transcription factor. Biophysical characterization of these MGs revealed the importance of optimized linker length, displaying a reduced entropic cost compared to the linear counterparts, while preserving key contacts with 14-3-3. Binding assays demonstrated that the macrocycles selectively and cooperatively stabilized the 14-3-3/ChREBP complex, with an intriguing inverse relationship between intrinsic binding affinity to 14-3-3 and cooperativity in PPI stabilization. Ternary co-crystal structures of the macrocycles binding at the composite 14-3-3/ChREBP interface provided a molecular rationale for the affinity and cooperativity differences. Overall, this study highlights structural, kinetic, and thermodynamic features that guide effective macrocyclic MG design and brings forward the crucial interplay of affinity and cooperativity in stabilizing PPIs.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, 5600MB, The Netherlands.
Organizational Affiliation: