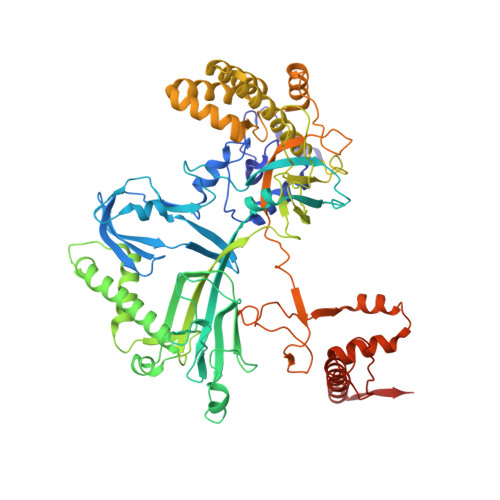
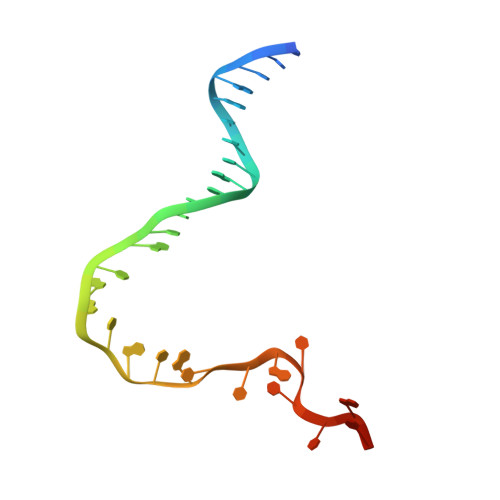
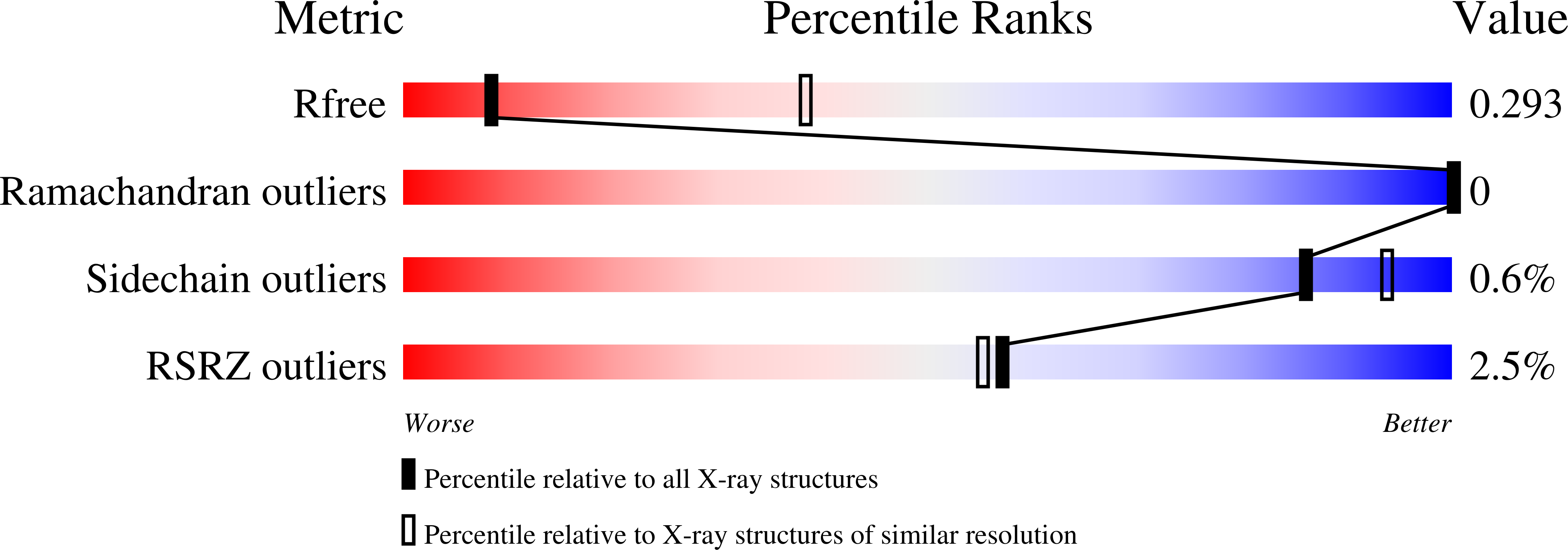Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Fraser, A., Sokolova, M.L., Drobysheva, A.V., Gordeeva, J.V., Borukhov, S., Jumper, J., Severinov, K.V., Leiman, P.G.(2022) Nat Commun 13: 3526-3526
- PubMed: 35725571
- DOI: https://doi.org/10.1038/s41467-022-31214-6
- Primary Citation of Related Structures:
7S00, 7S01, 7UM0, 7UM1 - PubMed Abstract:
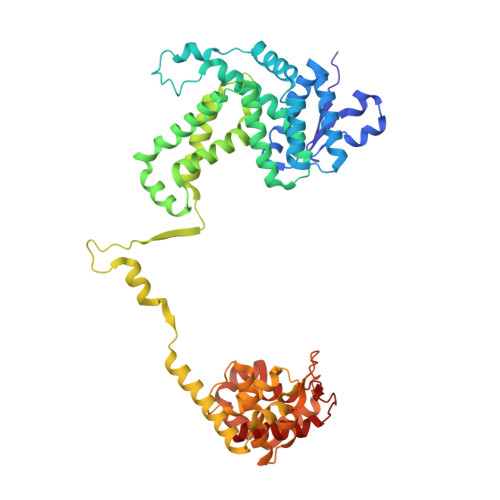
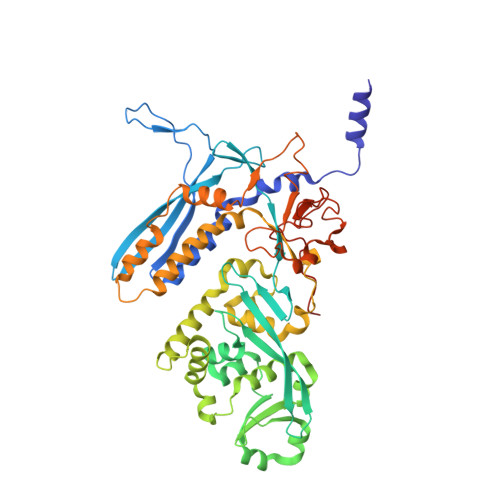
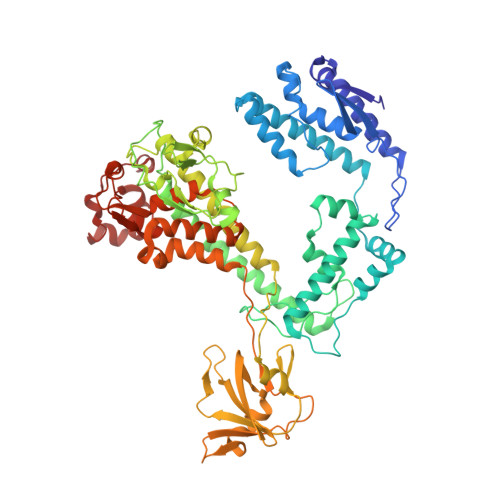
Recognition of promoters in bacterial RNA polymerases (RNAPs) is controlled by sigma subunits. The key sequence motif recognized by the sigma, the -10 promoter element, is located in the non-template strand of the double-stranded DNA molecule ~10 nucleotides upstream of the transcription start site. Here, we explain the mechanism by which the phage AR9 non-virion RNAP (nvRNAP), a bacterial RNAP homolog, recognizes the -10 element of its deoxyuridine-containing promoter in the template strand. The AR9 sigma-like subunit, the nvRNAP enzyme core, and the template strand together form two nucleotide base-accepting pockets whose shapes dictate the requirement for the conserved deoxyuridines. A single amino acid substitution in the AR9 sigma-like subunit allows one of these pockets to accept a thymine thus expanding the promoter consensus. Our work demonstrates the extent to which viruses can evolve host-derived multisubunit enzymes to make transcription of their own genes independent of the host.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, TX, 77555-0647, USA.