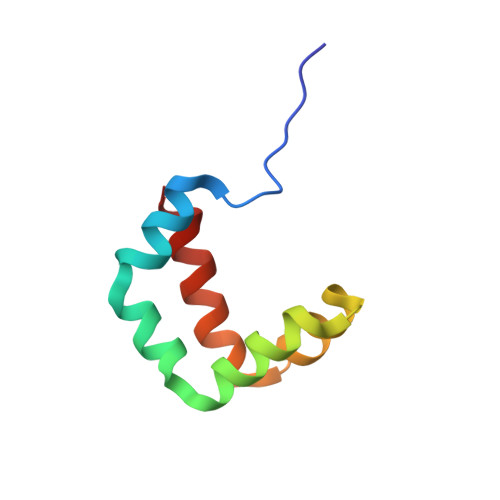
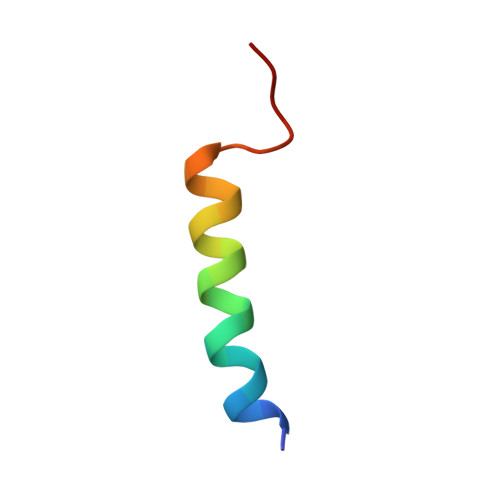
Structural Analysis of the Plasmodial Proteins ZNHIT3 and NUFIP1 Provides Insights into the Selectivity of a Conserved Interaction.
Chagot, M.E., Boutilliat, A., Kriznik, A., Quinternet, M.(2022) Biochemistry 61: 479-493
- PubMed: 35315277
- DOI: https://doi.org/10.1021/acs.biochem.1c00792
- Primary Citation of Related Structures:
7QDW - PubMed Abstract:
Malaria is a widespread and lethal disease caused by the Plasmodium parasites that can infect human beings through Anopheles mosquitoes. For that reason, the biology of Plasmodium needs to be studied to develop antimalarial treatments. By determining the three-dimensional structures of macromolecules, structural biology helps to understand the function of proteins and can reveal how interactions occur between biological partners. Here, we studied the ZNHIT3 and NUFIP1 proteins from Plasmodium falciparum , two proteins tightly linked to the ribosome biology. Due to their important functions in post-translational modifications of ribosomal RNAs and in ribophagy, these proteins participate in the survival of cells. In this study, we solved the three-dimensional structure of a thermally stable and species-dependent complex between fragments of these proteins. Our results were compared to the AlphaFold predictions, which motivated the study of the free ZNHIT3 fragment that binds NUFIP1. We showed that the latter fragment multimerized in vitro but also had the inner ability to change its conformation to escape the solvent exposition of key hydrophobic residues involved in the interaction with NUFIP1. Our data could open the gate to selective drug discovery processes involving these two proteins.
Organizational Affiliation:
Université de Lorraine, CNRS, IMoPA, F-54000 Nancy, France.