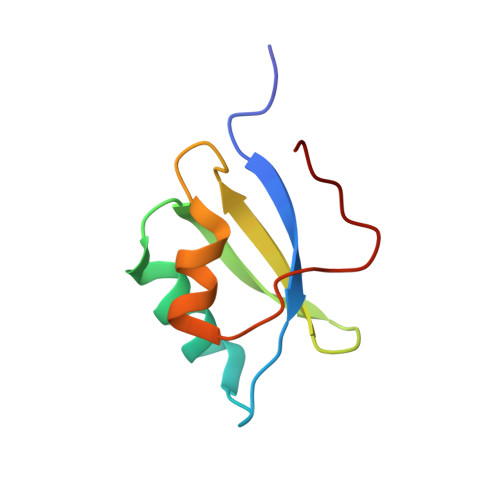
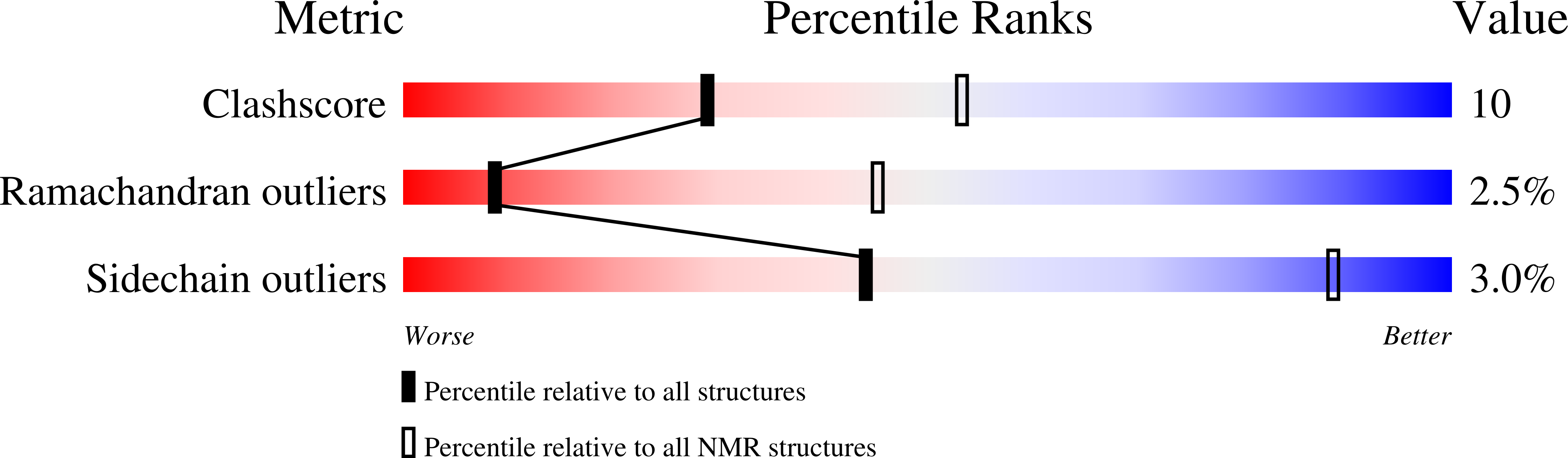At sixes and sevens: cryptic domain in the metal binding chain of the human copper transporter ATP7A.
Uhlemann, E.E., Lee, W., Tonelli, M., Dmitriev, O.Y.(2021) Biophys J 120: 4600-4607
- PubMed: 34461106
- DOI: https://doi.org/10.1016/j.bpj.2021.08.029
- Primary Citation of Related Structures:
7LU8 - PubMed Abstract:
ATP7A and ATP7B are structurally similar but functionally distinct active copper transporters that regulate copper levels in the human cells and deliver copper to the biosynthetic pathways. Both proteins have a chain of six cytosolic metal-binding domains (MBDs) believed to be involved in the copper-dependent regulation of the activity and intracellular localization of these enzymes. Although all the MBDs are quite similar in structure, their spacing differs markedly between ATP7A and ATP7B. We show by NMR that the long polypeptide between MBD1 and MBD2 of ATP7A forms an additional seventh metastable domain, which we called HMA1A (heavy metal associated domain 1A). The structure of HMA1A resembles the MBDs but contains no copper-binding site. The HMA1A domain, which is unique to ATP7A, may modulate regulatory interactions between MBD1-3, contributing to the distinct functional properties of ATP7A and ATP7B.
Organizational Affiliation:
Department of Biochemistry, Microbiology and Immunology, University of Saskatchewan, Saskatoon, Saskatchewan, Canada.