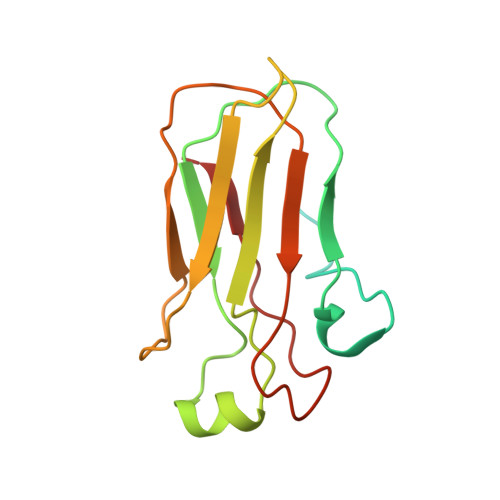
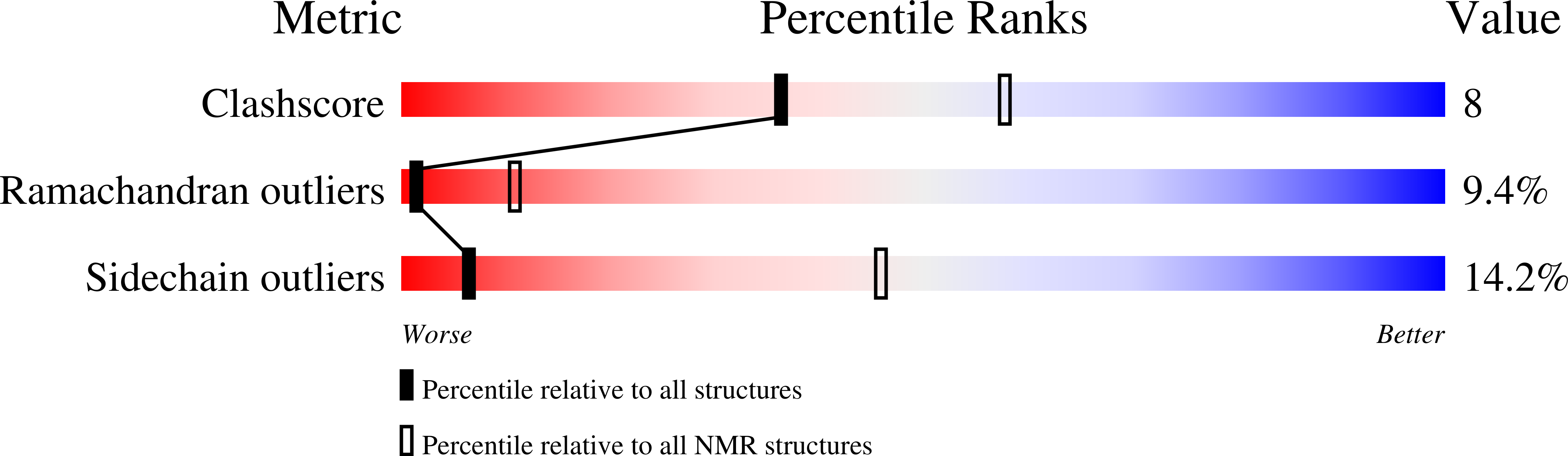Structure of the Streptococcus pyogenes NAD + Glycohydrolase Translocation Domain and Its Essential Role in Toxin Binding to Oropharyngeal Keratinocytes.
Velarde, J.J., Piai, A., Lichtenstein, I.J., Lynskey, N.N., Chou, J.J., Wessels, M.R.(2022) J Bacteriol 204: e0036621-e0036621
- PubMed: 34694903
- DOI: https://doi.org/10.1128/JB.00366-21
- Primary Citation of Related Structures:
7JI1 - PubMed Abstract:
The emergence and continued dominance of a Streptococcus pyogenes (group A Streptococcus, GAS) M1T1 clonal group is temporally correlated with acquisition of genomic sequences that confer high level expression of cotoxins streptolysin O (SLO) and NAD + -glycohydrolase (NADase). Experimental infection models have provided evidence that both toxins are important contributors to GAS virulence. SLO is a cholesterol-dependent pore-forming toxin capable of lysing virtually all types of mammalian cells. NADase, which is composed of an N-terminal translocation domain and C-terminal glycohydrolase domain, acts as an intracellular toxin that depletes host cell energy stores. NADase is dependent on SLO for internalization into epithelial cells, but its mechanism of interaction with the cell surface and details of its translocation mechanism remain unclear. In this study we found that NADase can bind oropharyngeal epithelial cells independently of SLO. This interaction is mediated by both domains of the toxin. We determined by NMR the structure of the translocation domain to be a β-sandwich with a disordered N-terminal region. The folded region of the domain has structural homology to carbohydrate binding modules. We show that excess NADase inhibits SLO-mediated hemolysis and binding to epithelial cells in vitro , suggesting NADase and SLO have shared surface receptors. This effect is abrogated by disruption of a putative carbohydrate binding site on the NADase translocation domain. Our data are consistent with a model whereby interactions of the NADase glycohydrolase domain and translocation domain with SLO and the cell surface increase avidity of NADase binding and facilitate toxin-toxin and toxin-cell surface interactions. IMPORTANCE NADase and streptolysin O (SLO) are secreted toxins important for pathogenesis of group A Streptococcus, the agent of strep throat and severe invasive infections. The two toxins interact in solution and mutually enhance cytotoxic activity. We now find that NADase is capable of binding to the surface of human cells independently of SLO. Structural analysis of the previously uncharacterized translocation domain of NADase suggests that it contains a carbohydrate binding module. The NADase translocation domain and SLO appear to recognize similar glycan structures on the cell surface, which may be one mechanism through which NADase enhances SLO pore-forming activity during infection. Our findings provide new insight into the NADase toxin and its functional interactions with SLO during streptococcal infection.
Organizational Affiliation:
Division of Infectious Diseases, Boston Children's Hospital, and Department of Pediatrics, Harvard Medical Schoolgrid.471403.5, Boston, Massachusetts, USA.