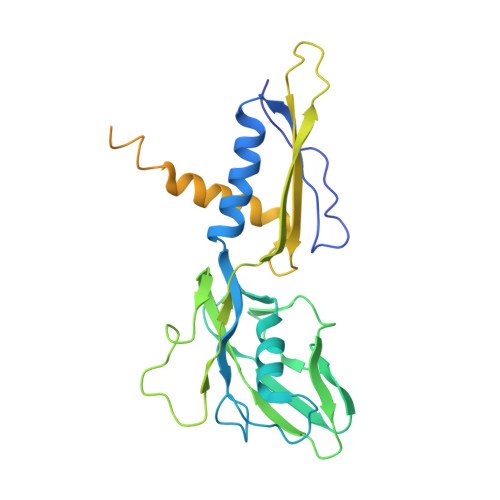
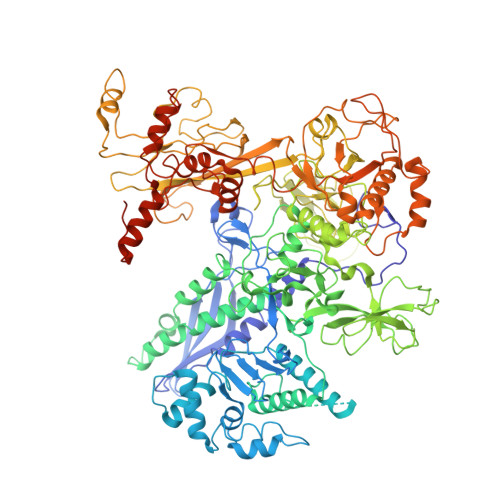
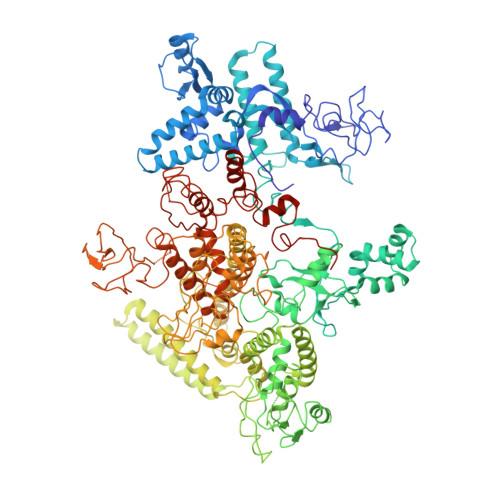
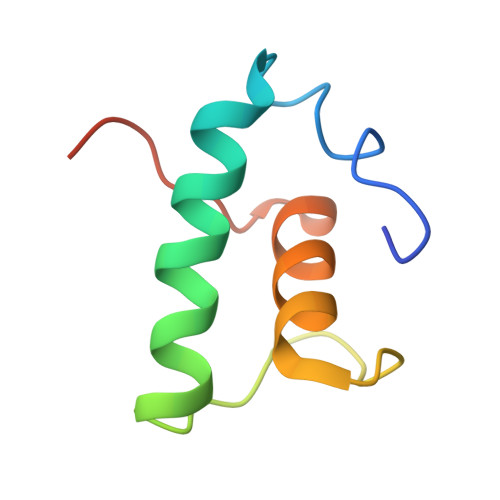
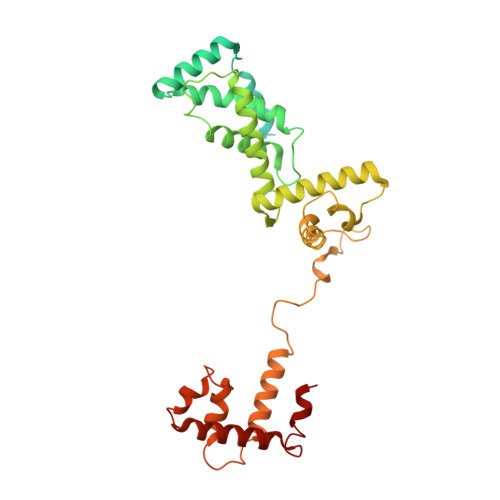
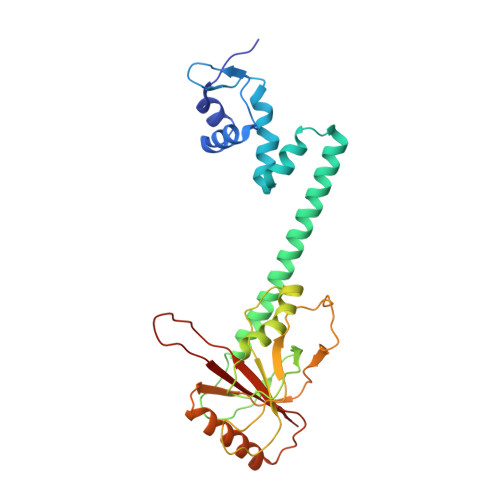
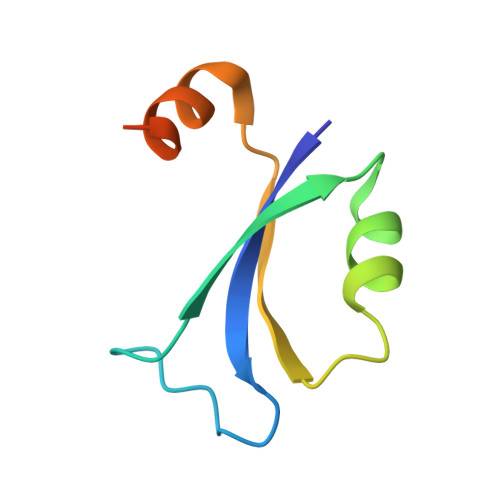
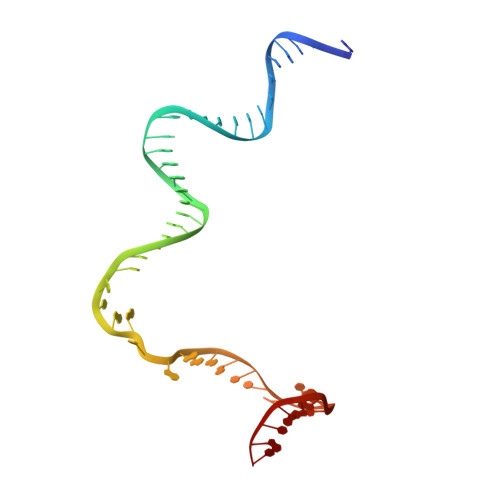
The bacterial multidrug resistance regulator BmrR distorts promoter DNA to activate transcription.
Fang, C., Li, L., Zhao, Y., Wu, X., Philips, S.J., You, L., Zhong, M., Shi, X., O'Halloran, T.V., Li, Q., Zhang, Y.(2020) Nat Commun 11: 6284-6284
- PubMed: 33293519
- DOI: https://doi.org/10.1038/s41467-020-20134-y
- Primary Citation of Related Structures:
7CKQ - PubMed Abstract:
The MerR-family proteins represent a unique family of bacteria transcription factors (TFs), which activate transcription in a manner distinct from canonical ones. Here, we report a cryo-EM structure of a B. subtilis transcription activation complex comprising B. subtilis six-subunit (2αββ'ωε) RNA Polymerase (RNAP) core enzyme, σ A , a promoter DNA, and the ligand-bound B. subtilis BmrR, a prototype of MerR-family TFs. The structure reveals that RNAP and BmrR recognize the upstream promoter DNA from opposite faces and induce four significant kinks from the -35 element to the -10 element of the promoter DNA in a cooperative manner, which restores otherwise inactive promoter activity by shortening the length of promoter non-optimal -35/-10 spacer. Our structure supports a DNA-distortion and RNAP-non-contact paradigm of transcriptional activation by MerR TFs.
Organizational Affiliation:
Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, 200032, Shanghai, China.