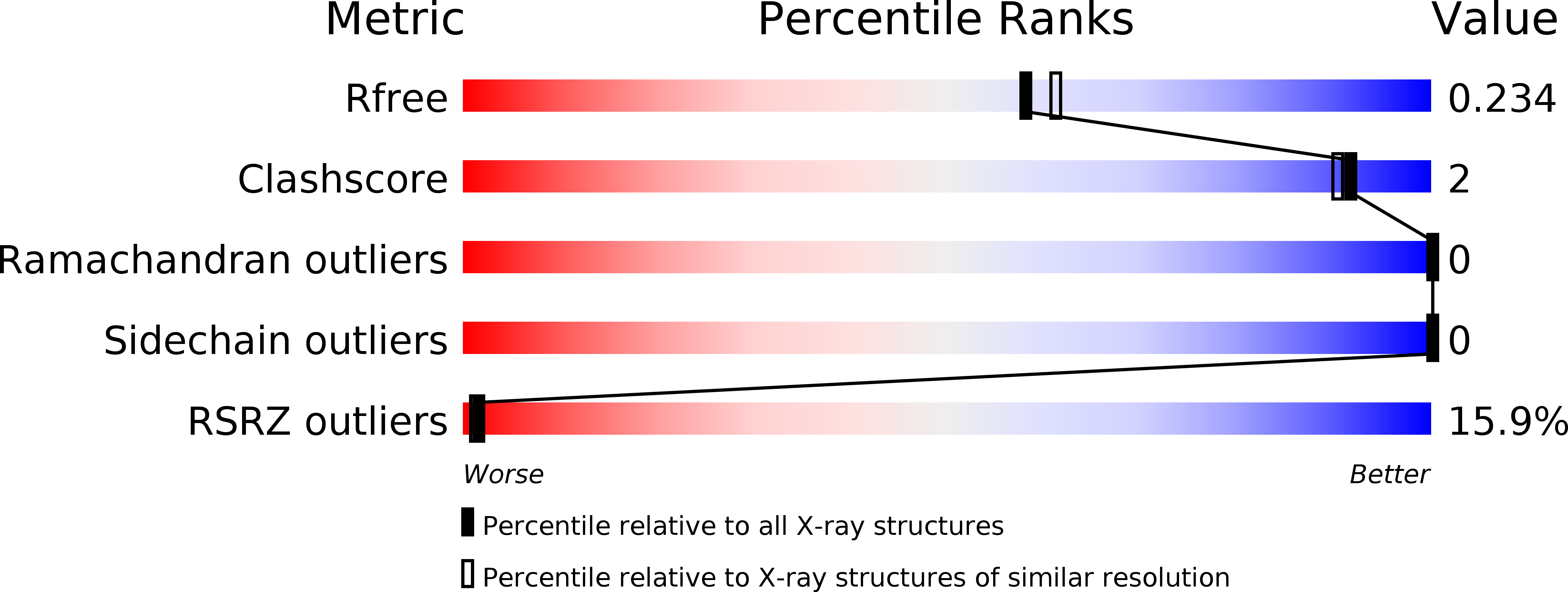
Crystal structures of the sheeppox virus encoded inhibitor of apoptosis SPPV14 bound to the proapoptotic BH3 peptides Hrk and Bax.
Suraweera, C.D., Burton, D.R., Hinds, M.G., Kvansakul, M.(2020) FEBS Lett 594: 2016-2026
- PubMed: 32390192
- DOI: https://doi.org/10.1002/1873-3468.13807
- Primary Citation of Related Structures:
6XY4, 6XY6 - PubMed Abstract:
Programmed death of infected cells is used by multicellular organisms to counter viral infections. Sheeppox virus encodes for SPPV14, a potent inhibitor of Bcl-2-mediated apoptosis. We reveal the structural basis of apoptosis inhibition by determining crystal structures of SPPV14 bound to BH3 motifs of proapoptotic Bax and Hrk. The structures show that SPPV14 engages BH3 peptides using the canonical ligand-binding groove. Unexpectedly, Arg84 from SPPV14 forms an ionic interaction with the conserved Asp in the BH3 motif in a manner that replaces the canonical ionic interaction seen in almost all host Bcl-2:BH3 motif complexes. These results reveal the flexibility of virus-encoded Bcl-2 proteins to mimic key interactions from endogenous host signalling pathways to retain BH3 binding and prosurvival functionality.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Vic., Australia.