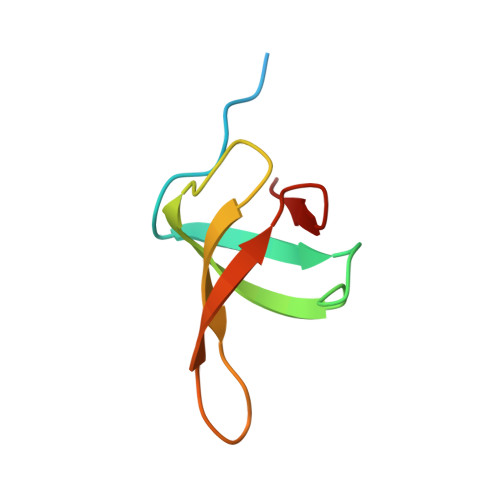
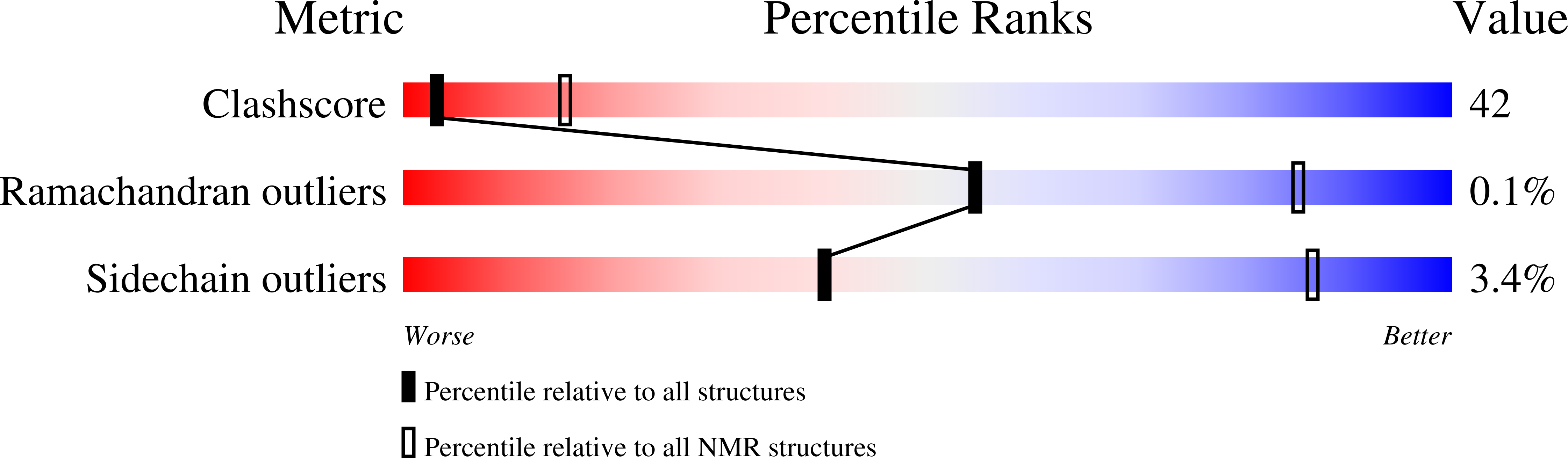Structural and thermodynamic analyses of the beta-to-alpha transformation in RfaH reveal principles of fold-switching proteins.
Zuber, P.K., Daviter, T., Heissmann, R., Persau, U., Schweimer, K., Knauer, S.H.(2022) Elife 11
- PubMed: 36255050
- DOI: https://doi.org/10.7554/eLife.76630
- Primary Citation of Related Structures:
6TF4 - PubMed Abstract:
The two-domain protein RfaH, a paralog of the universally conserved NusG/Spt5 transcription factors, is regulated by autoinhibition coupled to the reversible conformational switch of its 60-residue C-terminal Kyrpides, Ouzounis, Woese (KOW) domain between an α-hairpin and a β-barrel. In contrast, NusG/Spt5-KOW domains only occur in the β-barrel state. To understand the principles underlying the drastic fold switch in RfaH, we elucidated the thermodynamic stability and the structural dynamics of two RfaH- and four NusG/Spt5-KOW domains by combining biophysical and structural biology methods. We find that the RfaH-KOW β-barrel is thermodynamically less stable than that of most NusG/Spt5-KOWs and we show that it is in equilibrium with a globally unfolded species, which, strikingly, contains two helical regions that prime the transition toward the α-hairpin. Our results suggest that transiently structured elements in the unfolded conformation might drive the global folding transition in metamorphic proteins in general.
Organizational Affiliation:
Biochemistry IV - Biophysical Chemistry, University of Bayreuth, Bayreuth, Germany.