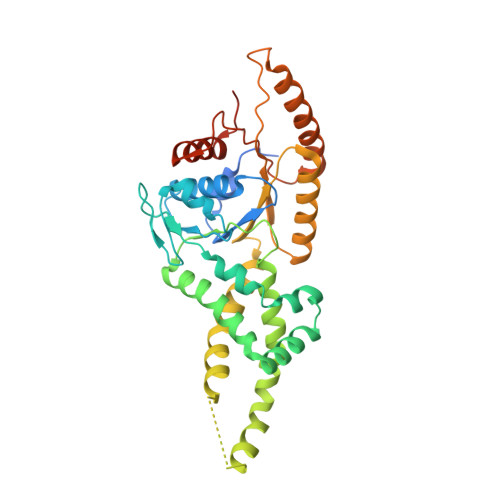
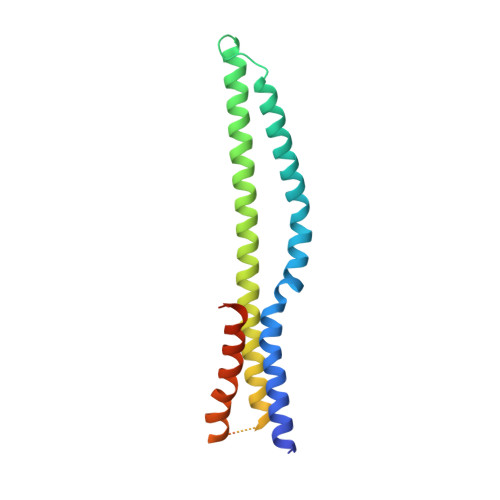
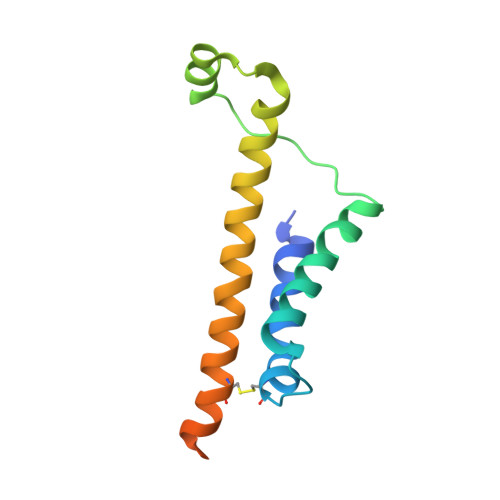
Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
McDowell, M.A., Heimes, M., Fiorentino, F., Mehmood, S., Farkas, A., Coy-Vergara, J., Wu, D., Bolla, J.R., Schmid, V., Heinze, R., Wild, K., Flemming, D., Pfeffer, S., Schwappach, B., Robinson, C.V., Sinning, I.(2020) Mol Cell 80: 72
- PubMed: 32910895
- DOI: https://doi.org/10.1016/j.molcel.2020.08.012
- Primary Citation of Related Structures:
6SO5 - PubMed Abstract:
Membrane protein biogenesis faces the challenge of chaperoning hydrophobic transmembrane helices for faithful membrane insertion. The guided entry of tail-anchored proteins (GET) pathway targets and inserts tail-anchored (TA) proteins into the endoplasmic reticulum (ER) membrane with an insertase (yeast Get1/Get2 or mammalian WRB/CAML) that captures the TA from a cytoplasmic chaperone (Get3 or TRC40, respectively). Here, we present cryo-electron microscopy reconstructions, native mass spectrometry, and structure-based mutagenesis of human WRB/CAML/TRC40 and yeast Get1/Get2/Get3 complexes. Get3 binding to the membrane insertase supports heterotetramer formation, and phosphatidylinositol binding at the heterotetramer interface stabilizes the insertase for efficient TA insertion in vivo. We identify a Get2/CAML cytoplasmic helix that forms a "gating" interaction with Get3/TRC40 important for TA insertion. Structural homology with YidC and the ER membrane protein complex (EMC) implicates an evolutionarily conserved insertion mechanism for divergent substrates utilizing a hydrophilic groove. Thus, we provide a detailed structural and mechanistic framework to understand TA membrane insertion.
Organizational Affiliation:
Heidelberg University Biochemistry Center (BZH), Im Neuenheimer Feld 328, 69120 Heidelberg, Germany. Electronic address: melanie.mcdowell@bzh.uni-heidelberg.de.