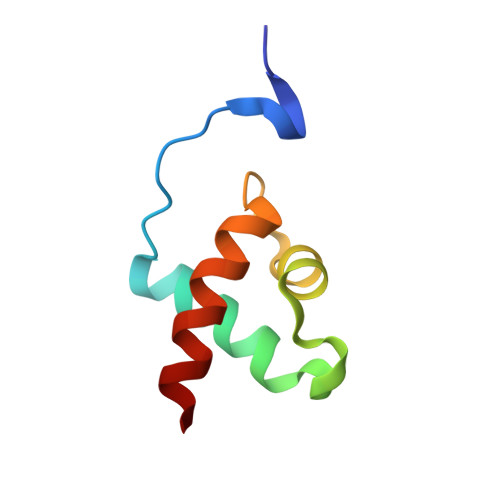
Molecular mechanisms of Evening Complex activity inArabidopsis.
Silva, C.S., Nayak, A., Lai, X., Hutin, S., Hugouvieux, V., Jung, J.H., Lopez-Vidriero, I., Franco-Zorrilla, J.M., Panigrahi, K.C.S., Nanao, M.H., Wigge, P.A., Zubieta, C.(2020) Proc Natl Acad Sci U S A 117: 6901-6909
- PubMed: 32165537
- DOI: https://doi.org/10.1073/pnas.1920972117
- Primary Citation of Related Structures:
6QEC - PubMed Abstract:
The Evening Complex (EC), composed of the DNA binding protein LUX ARRHYTHMO (LUX) and two additional proteins EARLY FLOWERING 3 (ELF3) and ELF4, is a transcriptional repressor complex and a core component of the plant circadian clock. In addition to maintaining oscillations in clock gene expression, the EC also participates in temperature and light entrainment, acting as an important environmental sensor and conveying this information to growth and developmental pathways. However, the molecular basis for EC DNA binding specificity and temperature-dependent activity were not known. Here, we solved the structure of the DNA binding domain of LUX in complex with DNA. Residues critical for high-affinity binding and direct base readout were determined and tested via site-directed mutagenesis in vitro and in vivo. Using extensive in vitro DNA binding assays of LUX alone and in complex with ELF3 and ELF4, we demonstrate that, while LUX alone binds DNA with high affinity, the LUX-ELF3 complex is a relatively poor binder of DNA. ELF4 restores binding to the complex. In vitro, the full EC is able to act as a direct thermosensor, with stronger DNA binding at 4 °C and weaker binding at 27 °C. In addition, an excess of ELF4 is able to restore EC binding even at 27 °C. Taken together, these data suggest that ELF4 is a key modulator of thermosensitive EC activity.
Organizational Affiliation:
Laboratoire de Physiologie Cellulaire and Végétale, Université Grenoble Alpes/Centre National de la Recherche Scientifique/Commissariat à l'Énergie Atomique et aux Énergies Alternatives/Institut National de la Recherche Agronomique/Interdisciplinary Research Institute of Grenoble, 38054 Grenoble, France; Csilva@embl.fr stephanie.hutin@univ-grenoble-alpes.fr Chloe.Zubieta@cea.fr.