Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Berg, J., Liu, J., Svahn, E., Ferguson-Miller, S., Brzezinski, P.(2019) Biochim Biophys Acta Bioenerg 1861: 148116-148116
- PubMed: 31733183
- DOI: https://doi.org/10.1016/j.bbabio.2019.148116
- Primary Citation of Related Structures:
6PW0, 6PW1 - PubMed Abstract:
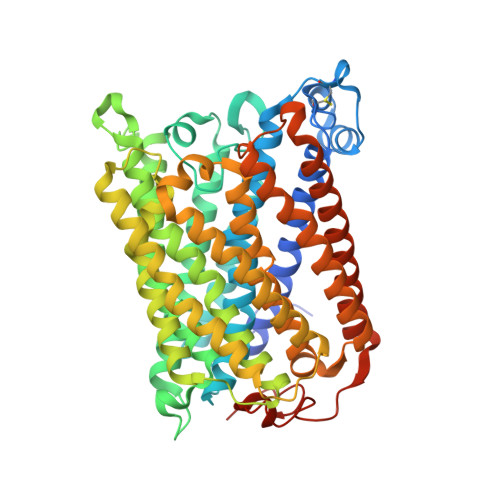
Data from earlier studies showed that minor structural changes at the surface of cytochrome c oxidase, in one of the proton-input pathways (the D pathway), result in dramatically decreased activity and a lower proton-pumping stoichiometry. To further investigate how changes around the D pathway orifice influence functionality of the enzyme, here we modified the nearby C-terminal loop of subunit I of the Rhodobacter sphaeroides cytochrome c oxidase. Removal of 16 residues from this flexible surface loop resulted in a decrease in the proton-pumping stoichiometry to <50% of that of the wild-type enzyme. Replacement of the protonatable residue Glu552, part of the same loop, by an Ala, resulted in a similar decrease in the proton-pumping stoichiometry without loss of the O 2 -reduction activity or changes in the proton-uptake kinetics. The data show that minor structural changes at the orifice of the D pathway, at a distance of ~40 Å from the proton gate of cytochrome c oxidase, may alter the proton-pumping stoichiometry of the enzyme.
Organizational Affiliation:
Department of Biochemistry and Biophysics, The Arrhenius Laboratories for Natural Sciences, Stockholm University, SE-106 91 Stockholm, Sweden.