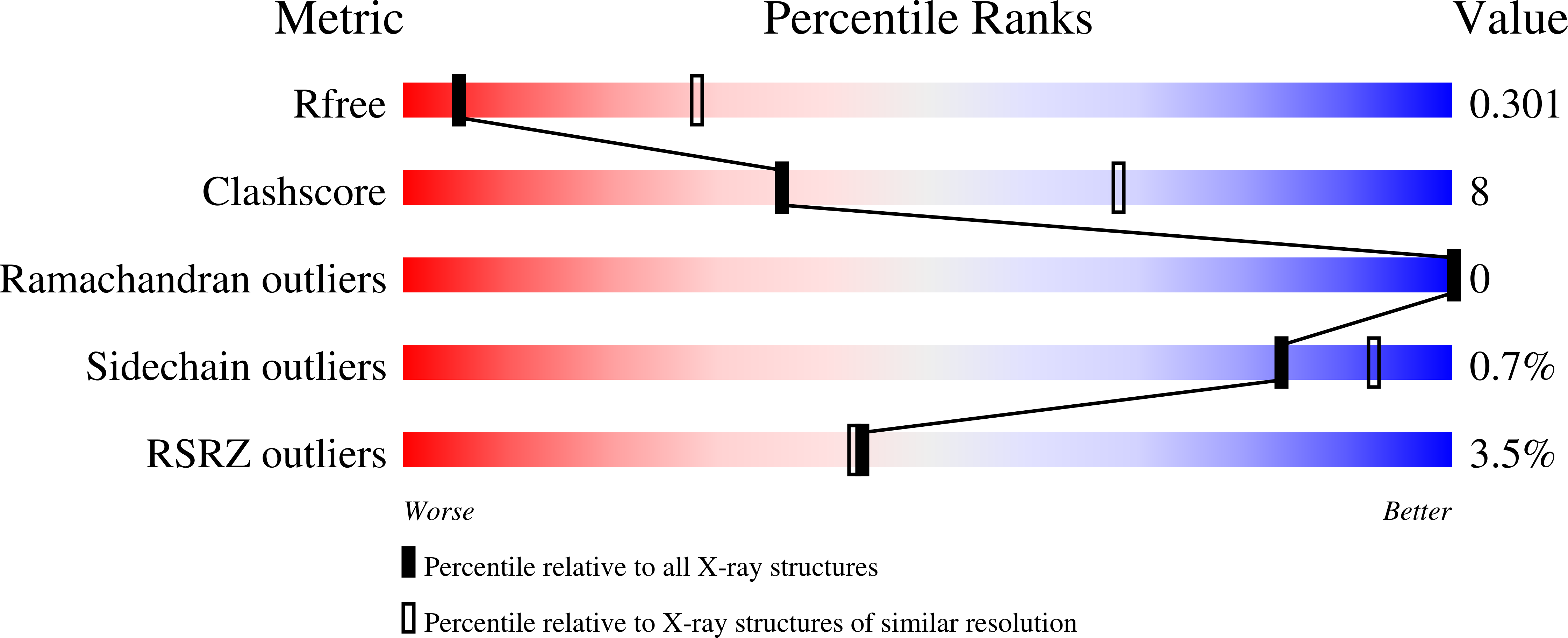
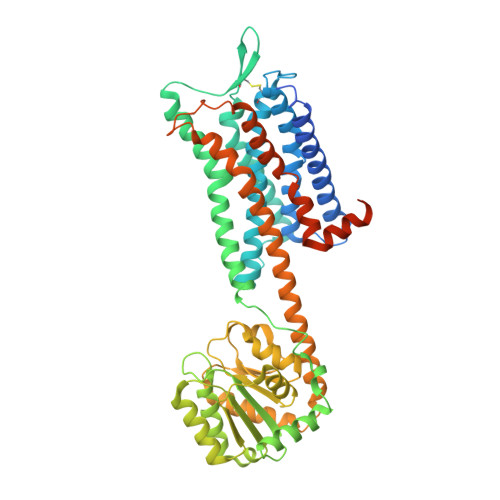
Crystal structure of the human NK1tachykinin receptor.
Yin, J., Chapman, K., Clark, L.D., Shao, Z., Borek, D., Xu, Q., Wang, J., Rosenbaum, D.M.(2018) Proc Natl Acad Sci U S A 115: 13264-13269
- PubMed: 30538204
- DOI: https://doi.org/10.1073/pnas.1812717115
- Primary Citation of Related Structures:
6E59 - PubMed Abstract:
The NK 1 tachykinin G-protein-coupled receptor (GPCR) binds substance P, the first neuropeptide to be discovered in mammals. Through activation of NK 1 R, substance P modulates a wide variety of physiological and disease processes including nociception, inflammation, and depression. Human NK 1 R (hNK 1 R) modulators have shown promise in clinical trials for migraine, depression, and emesis. However, the only currently approved drugs targeting hNK 1 R are inhibitors for chemotherapy-induced nausea and vomiting (CINV). To better understand the molecular basis of ligand recognition and selectivity, we solved the crystal structure of hNK 1 R bound to the inhibitor L760735, a close analog of the drug aprepitant. Our crystal structure reveals the basis for antagonist interaction in the deep and narrow orthosteric pocket of the receptor. We used our structure as a template for computational docking and molecular-dynamics simulations to dissect the energetic importance of binding pocket interactions and model the binding of aprepitant. The structure of hNK 1 R is a valuable tool in the further development of tachykinin receptor modulators for multiple clinical applications.
Organizational Affiliation:
Department of Biophysics, The University of Texas Southwestern Medical Center, Dallas, TX 75390.