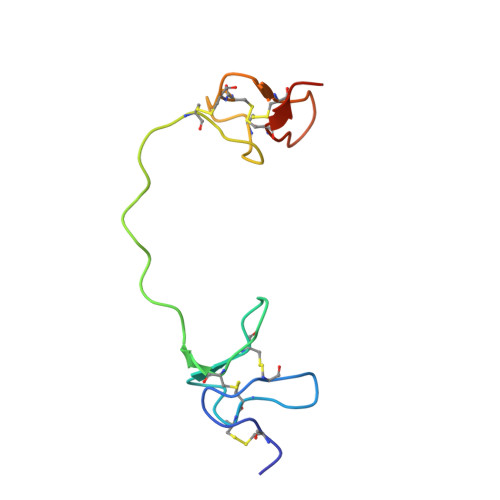
Structural Basis of the Bivalency of the TRPV1 Agonist DkTx.
Ramanujam, V., Crawford, T., Cristofori-Armstrong, B., Deuis, J.R., Jia, X., Maxwell, M.J., Jami, S., Ma, L., Vetter, I., Mobli, M.(2024) Angew Chem Int Ed Engl 63: e202314621-e202314621
- PubMed: 37953402
- DOI: https://doi.org/10.1002/anie.202314621
- Primary Citation of Related Structures:
6CUC - PubMed Abstract:
Bivalency is a prevalent natural mechanism to enhance receptor avidity. Various two-domain disulfide-rich peptides exhibiting bivalent action have been identified from animal venoms. A unique characteristic of these peptides is that they induce a pharmacological response different from that provoked by any of the constituent domains. The enhanced potency and avidity of such peptides is therefore a consequence of their domain fusion by a peptide linker. The role of the linker itself, beyond conjugation, remains unclear. Here, we investigate how the linker affects the bivalency of the capsaicin receptor (TRPV1) agonist DkTx. We recombinantly produced isotope labelled DkTx using a protein splicing approach, to solve the high-resolution solution structure of DkTx, revealing residual linker order stabilised by linker-domain interactions leading to biased domain orientations. The significance of this was studied using a combination of mutagenesis, spin relaxation studies and electrophysiology measurements. Our results reveal that disrupting the pre-organisation of the domains of DkTx is accompanied by reductions in potency and onset of avidity. Our findings support a model of pre-configured two-domain binding, in favour of the previously suggested sequential binding model. This highlights the significance of ordered elements in linker design and the natural evolution of these in bivalent toxins.
Organizational Affiliation:
Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, St Lucia, 4072, Queensland, Australia.