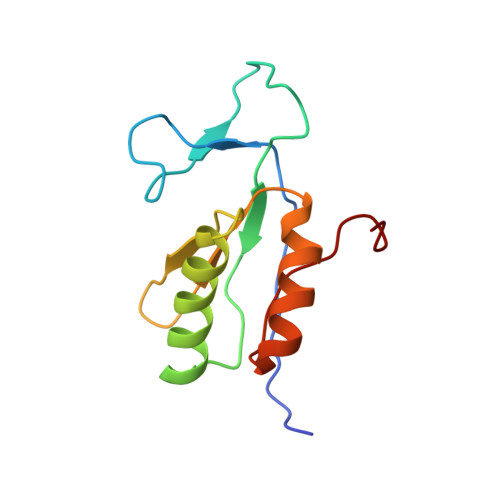
The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Mohanty, B., Hanson-Manful, P., Finn, T.J., Chambers, C.R., McKellar, J.L.O., Macindoe, I., Helder, S., Setiyaputra, S., Zhong, Y., Mackay, J.P., Patrick, W.M.(2019) Proteins 87: 699-705
- PubMed: 30958578
- DOI: https://doi.org/10.1002/prot.25687
- Primary Citation of Related Structures:
6BI6 - PubMed Abstract:
InterPro family IPR020489 comprises ~1000 uncharacterized bacterial proteins. Previously we showed that overexpressing the Escherichia coli representative of this family, EcYejG, conferred low-level resistance to aminoglycoside antibiotics. In an attempt to shed light on the biochemical function of EcYejG, we have solved its structure using multinuclear solution NMR spectroscopy. The structure most closely resembles that of domain III from elongation factor G (EF-G). EF-G catalyzes ribosomal translocation and mutations in EF-G have also been associated with aminoglycoside resistance. While we were unable to demonstrate a direct interaction between EcYejG and the ribosome, the protein might play a role in translation.
Organizational Affiliation:
Faculty of Pharmacy and Pharmaceutical Sciences, Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Australia.