Discovery of a novel azetidine scaffold for colony stimulating factor-1 receptor (CSF-1R) Type II inhibitors by the use of docking models.
Ikegashira, K., Ikenogami, T., Yamasaki, T., Hase, Y., Yamaguchi, T., Inagaki, K., Doi, S., Adachi, T., Koga, Y., Hashimoto, H.(2019) Bioorg Med Chem Lett 29: 115-118
- PubMed: 30442420
- DOI: https://doi.org/10.1016/j.bmcl.2018.10.051
- Primary Citation of Related Structures:
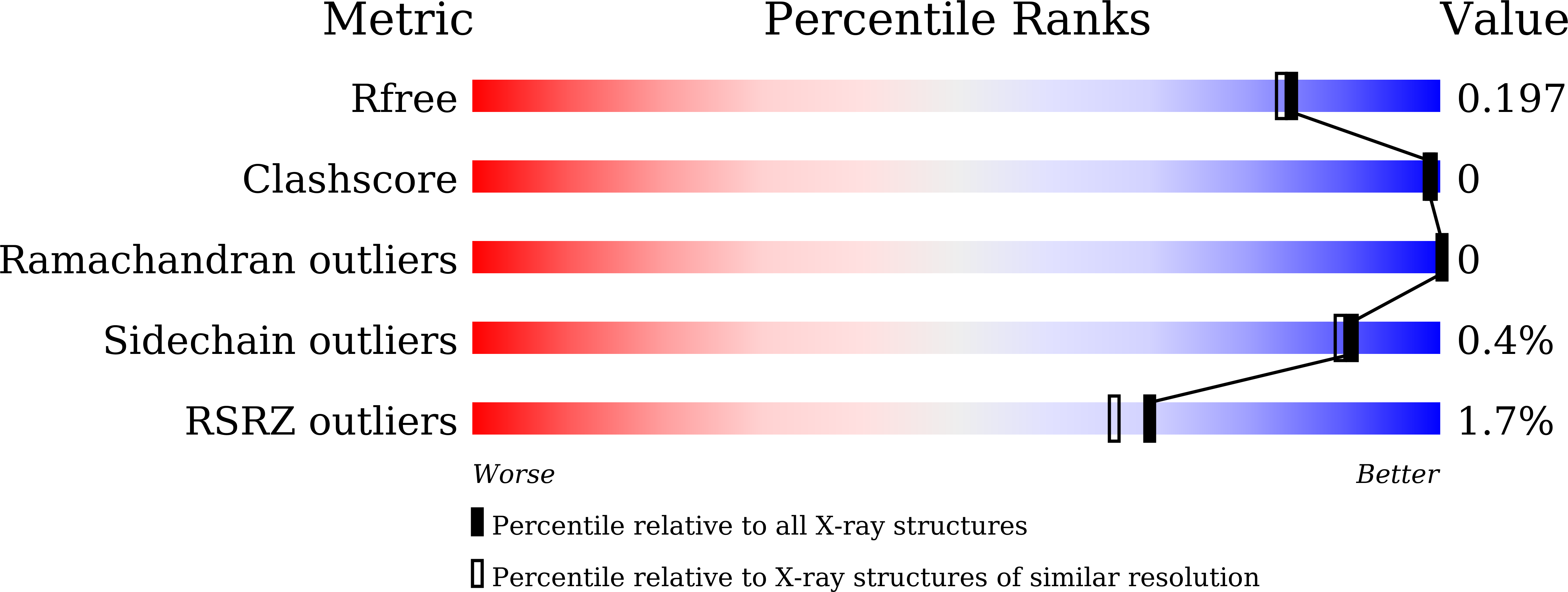
6IG8 - PubMed Abstract:
We report the discovery of a novel azetidine scaffold for colony stimulating factor-1 receptor (CSF-1R) Type II inhibitors by using a structure-based drug design (SBDD) based on a docking model. The work leads to the representative compound 4a with high CSF-1R inhibitory activity (IC 50 = 9.1 nM). The obtained crystal structure of an azetidine compound with CSF-1R, which matched our predicted docking model, demonstrates that the azetidine compounds bind to the DFG-out conformation of the protein as a Type II inhibitor.
Organizational Affiliation:
Central Pharmaceutical Research Institute, Japan Tobacco Inc., 1-1, Murasaki-cho, Takatsuki, Osaka 569-1125, Japan.