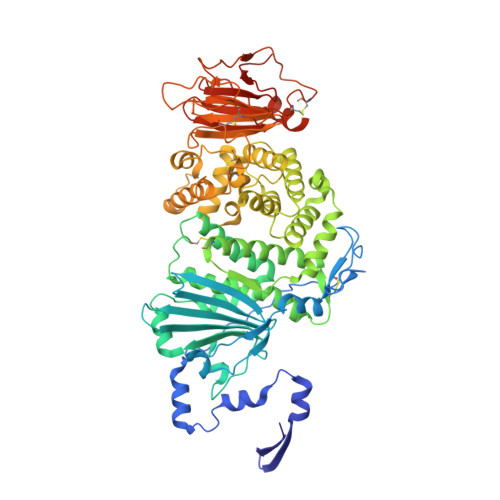
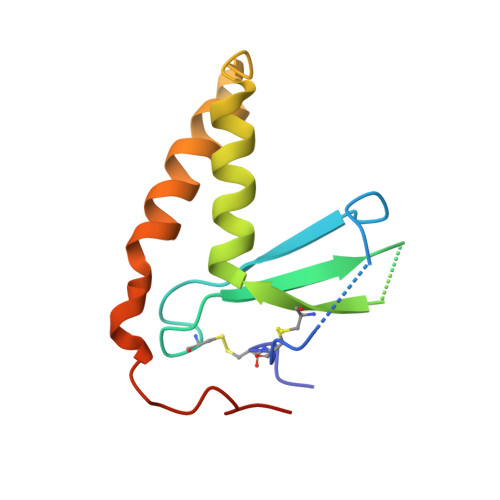
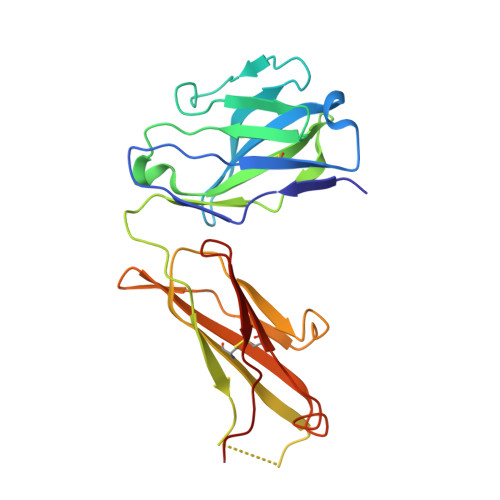
Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies.
Sathiyamoorthy, K., Jiang, J., Mohl, B.S., Chen, J., Zhou, Z.H., Longnecker, R., Jardetzky, T.S.(2017) Proc Natl Acad Sci U S A 114: E8703-E8710
- PubMed: 28939750
- DOI: https://doi.org/10.1073/pnas.1704661114
- Primary Citation of Related Structures:
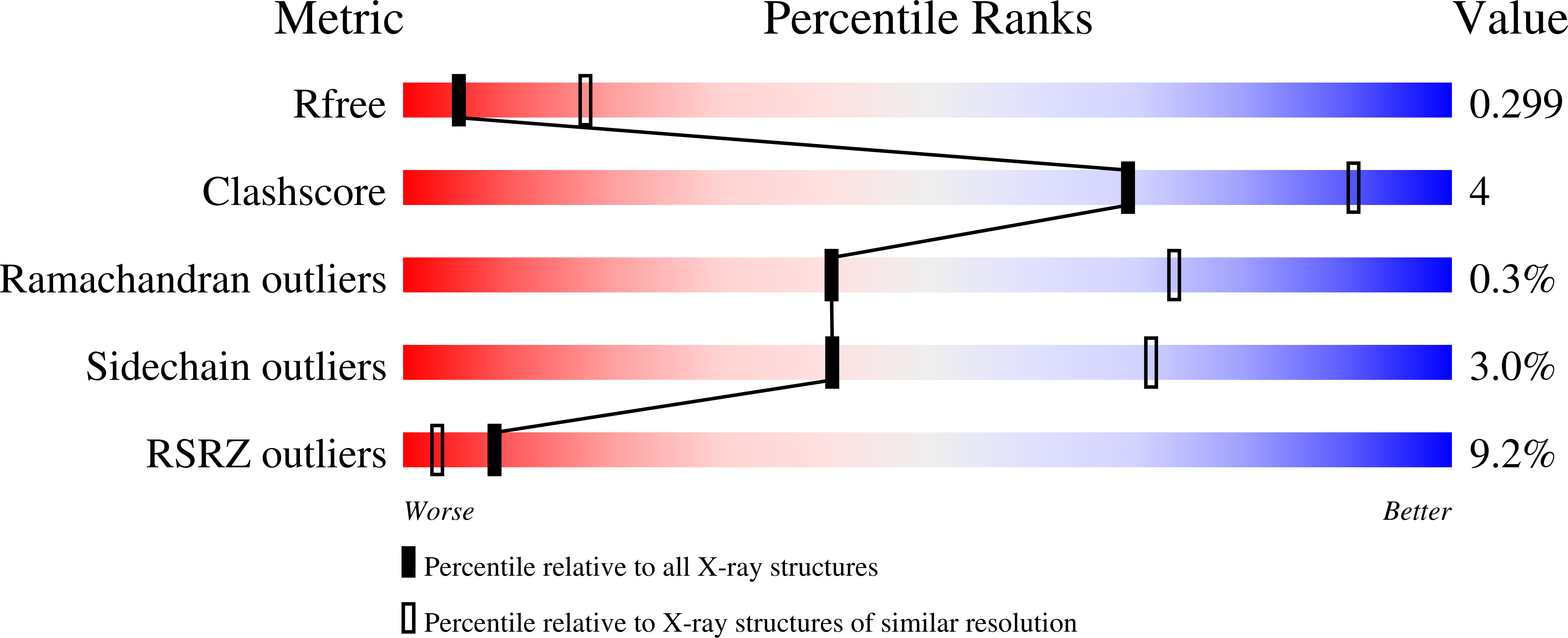
5W0K - PubMed Abstract:
Herpesvirus entry into cells requires the coordinated action of multiple virus envelope glycoproteins, including gH, gL, and gB. For EBV, the gp42 protein assembles into complexes with gHgL heterodimers and binds HLA class II to activate gB-mediated membrane fusion with B cells. EBV tropism is dictated by gp42 levels in the virion, as it inhibits entry into epithelial cells while promoting entry into B cells. The gHgL and gB proteins are targets of neutralizing antibodies and potential candidates for subunit vaccine development, but our understanding of their neutralizing epitopes and the mechanisms of inhibition remain relatively unexplored. Here we studied the structures and mechanisms of two anti-gHgL antibodies, CL40 and CL59, that block membrane fusion with both B cells and epithelial cells. We determined the structures of the CL40 and CL59 complexes with gHgL using X-ray crystallography and EM to identify their epitope locations. CL59 binds to the C-terminal domain IV of gH, while CL40 binds to a site occupied by the gp42 receptor binding domain. CL40 binding to gHgL/gp42 complexes is not blocked by gp42 and does not interfere with gp42 binding to HLA class II, indicating that its ability to block membrane fusion with B cells represents a defect in gB activation. These data indicate that anti-gHgL neutralizing antibodies can block gHgL-mediated activation of gB through different surface epitopes and mechanisms.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305.